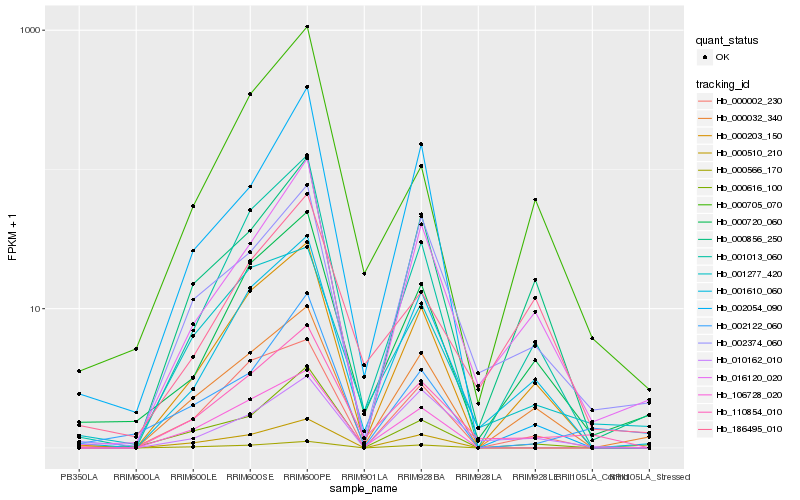
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_110854_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001610_060 |
0.1430552238 |
- |
- |
hypothetical protein TRIUR3_12067 [Triticum urartu] |
| 3 |
Hb_001277_420 |
0.1562729603 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 4 |
Hb_001013_060 |
0.1577067324 |
- |
- |
- |
| 5 |
Hb_000203_150 |
0.15963942 |
- |
- |
PREDICTED: uncharacterized protein LOC105116038 [Populus euphratica] |
| 6 |
Hb_016120_020 |
0.1612125204 |
- |
- |
PREDICTED: tetrahydrocannabinolic acid synthase-like [Jatropha curcas] |
| 7 |
Hb_000720_060 |
0.1638209017 |
- |
- |
PREDICTED: BON1-associated protein 2-like [Jatropha curcas] |
| 8 |
Hb_002374_060 |
0.1670427611 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 9 |
Hb_106728_020 |
0.1707208349 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000032_340 |
0.1858825936 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 11 |
Hb_000510_210 |
0.1866302037 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000002_230 |
0.1890656767 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 13 |
Hb_000856_250 |
0.1899663661 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_000616_100 |
0.1904800331 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000566_170 |
0.1926467305 |
transcription factor |
TF Family: MIKC |
PREDICTED: agamous-like MADS-box protein AGL9 homolog isoform X4 [Jatropha curcas] |
| 16 |
Hb_000705_070 |
0.1928805826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002122_060 |
0.1939245034 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_186495_010 |
0.1986020559 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 19 |
Hb_010162_010 |
0.1994818494 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 20 |
Hb_002054_090 |
0.1997255484 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Gossypium raimondii] |