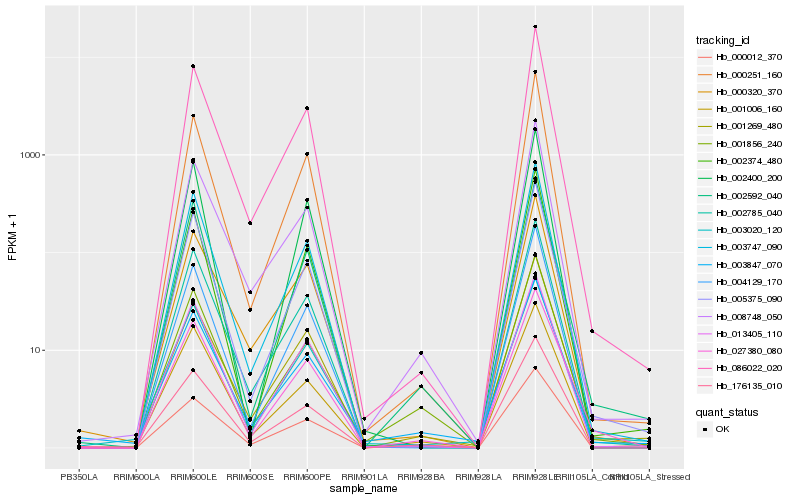
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 2 |
Hb_005375_090 |
0.0596719729 |
- |
- |
PREDICTED: protein CURVATURE THYLAKOID 1B, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002785_040 |
0.0665621725 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_003747_090 |
0.0669540027 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 5 |
Hb_002592_040 |
0.0684698288 |
- |
- |
ACC oxidase 3 [Hevea brasiliensis] |
| 6 |
Hb_013405_110 |
0.0703893426 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 7 |
Hb_086022_020 |
0.0711688513 |
- |
- |
RecName: Full=Ribulose bisphosphate carboxylase small chain, chloroplastic; Short=RuBisCO small subunit; Flags: Precursor [Hevea brasiliensis] |
| 8 |
Hb_001856_240 |
0.0756912779 |
transcription factor |
TF Family: TCP |
Plastid transcription factor 1, putative isoform 1 [Theobroma cacao] |
| 9 |
Hb_003020_120 |
0.077052196 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 10 |
Hb_000012_370 |
0.0780865216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001006_160 |
0.0785079778 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_008748_050 |
0.0785369449 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_004129_170 |
0.0790280327 |
- |
- |
PREDICTED: protein trichome birefringence-like 43 [Jatropha curcas] |
| 14 |
Hb_176135_010 |
0.0801297192 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 15 |
Hb_000251_160 |
0.082814904 |
- |
- |
chlorophyll A/B binding protein, putative [Ricinus communis] |
| 16 |
Hb_002374_480 |
0.0828550599 |
- |
- |
Photosystem II 22 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_003847_070 |
0.0829423215 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000320_370 |
0.0834643198 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002400_200 |
0.0847817162 |
- |
- |
14 kDa proline-rich protein DC2.15 precursor, putative [Ricinus communis] |
| 20 |
Hb_001269_480 |
0.0855412755 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 1, chloroplastic [Jatropha curcas] |