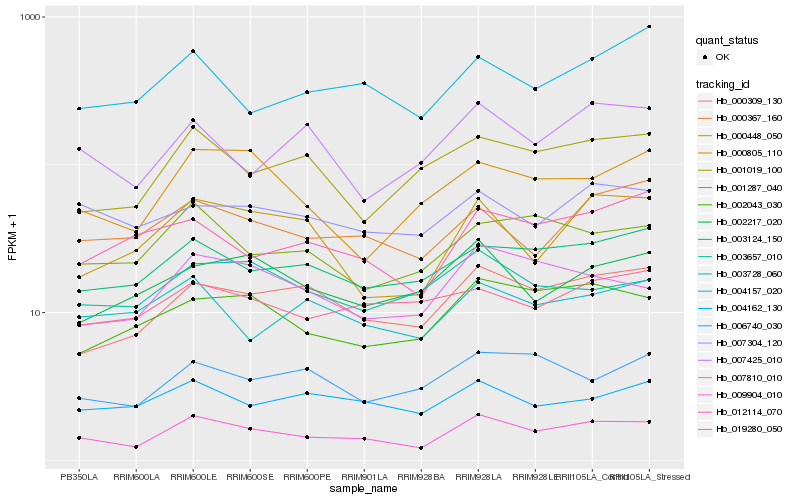
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009904_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_16885 [Jatropha curcas] |
| 2 |
Hb_000309_130 |
0.1137964226 |
- |
- |
hypothetical protein JCGZ_09592 [Jatropha curcas] |
| 3 |
Hb_007810_010 |
0.1147652132 |
- |
- |
PREDICTED: uncharacterized protein LOC105647655 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003124_150 |
0.1164487522 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 5 |
Hb_002043_030 |
0.1209969301 |
- |
- |
hypothetical protein RCOM_1486170 [Ricinus communis] |
| 6 |
Hb_000367_160 |
0.12361517 |
- |
- |
PREDICTED: SNAP25 homologous protein SNAP33 [Populus euphratica] |
| 7 |
Hb_001287_040 |
0.1245615541 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 8 |
Hb_004162_130 |
0.1279900361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007304_120 |
0.1318231079 |
- |
- |
NAD(P)-binding Rossmann-fold superfamily protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_000448_050 |
0.1325880294 |
- |
- |
PREDICTED: ubiquinol oxidase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000805_110 |
0.1326352057 |
- |
- |
PREDICTED: uncharacterized protein C24B11.05 [Jatropha curcas] |
| 12 |
Hb_001019_100 |
0.1327973187 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 13 |
Hb_003728_060 |
0.1337760647 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 14 |
Hb_003657_010 |
0.1344980638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002217_020 |
0.13516838 |
- |
- |
PREDICTED: uncharacterized protein LOC105643601 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004157_020 |
0.1354472983 |
- |
- |
PREDICTED: histone H2B-like [Gossypium raimondii] |
| 17 |
Hb_012114_070 |
0.1365683842 |
- |
- |
PREDICTED: uncharacterized protein LOC105645812 [Jatropha curcas] |
| 18 |
Hb_007425_010 |
0.1388658253 |
- |
- |
uncharacterized protein LOC100499741 [Glycine max] |
| 19 |
Hb_006740_030 |
0.1389400936 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_019280_050 |
0.1393652569 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 2 [Jatropha curcas] |