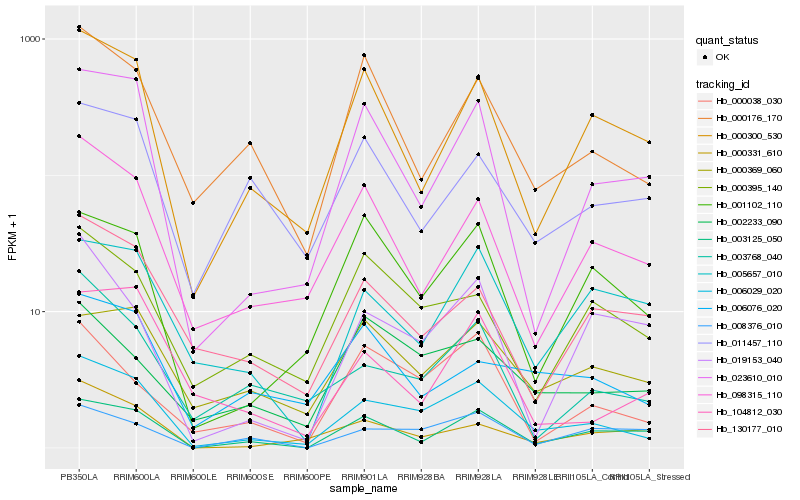
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006029_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1, partial [Vitis vinifera] |
| 2 |
Hb_023610_010 |
0.1783576503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000300_530 |
0.1795474307 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0286299-like [Gossypium raimondii] |
| 4 |
Hb_098315_110 |
0.1878226199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000176_170 |
0.1905852509 |
- |
- |
PREDICTED: tetratricopeptide repeat protein 1 [Jatropha curcas] |
| 6 |
Hb_003768_040 |
0.1928963486 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_003125_050 |
0.1953346325 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001102_110 |
0.1975877755 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1-like [Jatropha curcas] |
| 9 |
Hb_000038_030 |
0.2023669612 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 7-like [Glycine max] |
| 10 |
Hb_000369_060 |
0.202636691 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 11 |
Hb_130177_010 |
0.2046195827 |
- |
- |
hypothetical protein RCOM_0897890 [Ricinus communis] |
| 12 |
Hb_000331_610 |
0.2060876031 |
- |
- |
hypothetical protein B456_003G108600 [Gossypium raimondii] |
| 13 |
Hb_000395_140 |
0.2063840279 |
- |
- |
Oligopeptide transporter, putative [Ricinus communis] |
| 14 |
Hb_002233_090 |
0.2073264749 |
- |
- |
PREDICTED: bromodomain-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_006076_020 |
0.2088537894 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Vitis vinifera] |
| 16 |
Hb_005657_010 |
0.212043986 |
- |
- |
- |
| 17 |
Hb_104812_030 |
0.2151856283 |
- |
- |
hypothetical protein POPTR_0178s00210g [Populus trichocarpa] |
| 18 |
Hb_011457_110 |
0.2198049659 |
- |
- |
PREDICTED: protein NETWORKED 1D [Jatropha curcas] |
| 19 |
Hb_008376_010 |
0.2222889497 |
- |
- |
- |
| 20 |
Hb_019153_040 |
0.2222945012 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
putative 1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |