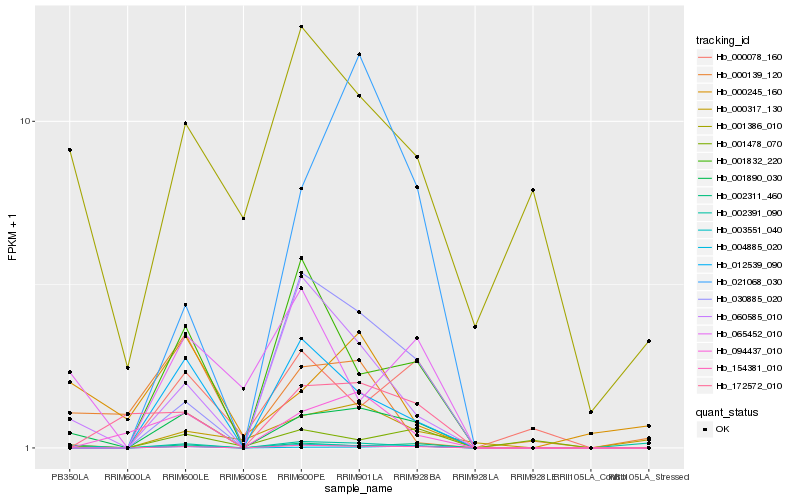
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001890_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_154381_010 |
0.2598676261 |
- |
- |
- |
| 3 |
Hb_003551_040 |
0.260261541 |
- |
- |
PREDICTED: uncharacterized protein LOC103331076 [Prunus mume] |
| 4 |
Hb_060585_010 |
0.2916870545 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_002311_460 |
0.295046901 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 6 |
Hb_012539_090 |
0.3125079741 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: U-box domain-containing protein 26, partial [Eucalyptus grandis] |
| 7 |
Hb_004885_020 |
0.313871046 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_065452_010 |
0.3269705606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001832_220 |
0.3316310362 |
- |
- |
- |
| 10 |
Hb_094437_010 |
0.3343728611 |
- |
- |
- |
| 11 |
Hb_000317_130 |
0.3367229112 |
- |
- |
vacuolar membrane protein pep3, putative [Ricinus communis] |
| 12 |
Hb_030885_020 |
0.3368316768 |
- |
- |
- |
| 13 |
Hb_000245_160 |
0.3381811223 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 14 |
Hb_001478_070 |
0.34541311 |
- |
- |
PREDICTED: uncharacterized protein LOC105789550 [Gossypium raimondii] |
| 15 |
Hb_021068_030 |
0.3482856052 |
transcription factor |
TF Family: M-type |
hypothetical protein B456_010G032700 [Gossypium raimondii] |
| 16 |
Hb_002391_090 |
0.3490621073 |
- |
- |
PREDICTED: IST1-like protein [Jatropha curcas] |
| 17 |
Hb_172572_010 |
0.3535426639 |
- |
- |
PREDICTED: ubiquitin thioesterase otubain-like [Jatropha curcas] |
| 18 |
Hb_000139_120 |
0.3541660441 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 19 |
Hb_001386_010 |
0.3563020444 |
- |
- |
PREDICTED: uncharacterized protein LOC105632444 [Jatropha curcas] |
| 20 |
Hb_000078_160 |
0.3591475041 |
- |
- |
PREDICTED: protein LURP-one-related 12-like [Jatropha curcas] |