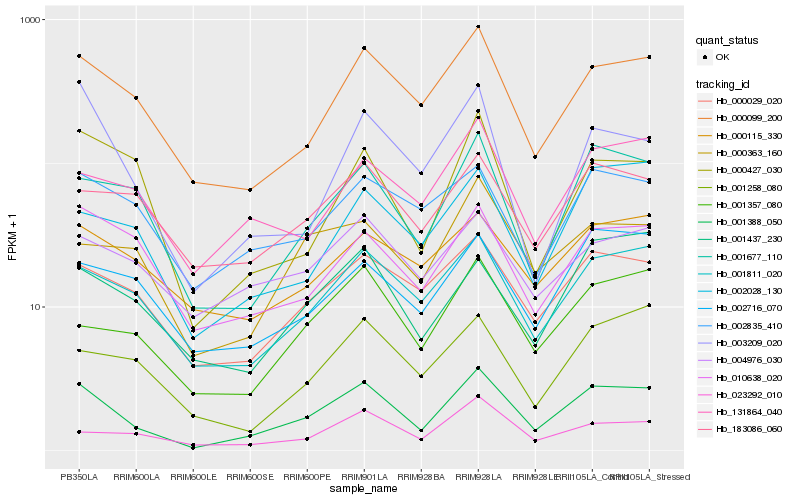
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001388_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_001811_020 |
0.1267629923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000099_200 |
0.1280715004 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001677_110 |
0.1354404872 |
- |
- |
Protein YIPF1, putative [Ricinus communis] |
| 5 |
Hb_000029_020 |
0.1395817043 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 6 |
Hb_001357_080 |
0.1465967443 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 7 |
Hb_001437_230 |
0.1576004689 |
- |
- |
PREDICTED: uncharacterized protein LOC105628035 [Jatropha curcas] |
| 8 |
Hb_131864_040 |
0.1577037022 |
- |
- |
PREDICTED: uncharacterized protein LOC105649406 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002028_130 |
0.1592400419 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 10 |
Hb_023292_010 |
0.1592479148 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 11 |
Hb_002716_070 |
0.1592809811 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 12 |
Hb_010638_020 |
0.1597812588 |
- |
- |
PREDICTED: uncharacterized protein LOC105638189 isoform X3 [Jatropha curcas] |
| 13 |
Hb_183086_060 |
0.1625262301 |
desease resistance |
Gene Name: AAA |
PREDICTED: ATPase family AAA domain-containing protein 1-B [Jatropha curcas] |
| 14 |
Hb_003209_020 |
0.1625283237 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 10 [Jatropha curcas] |
| 15 |
Hb_000363_160 |
0.1625439984 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 16 |
Hb_001258_080 |
0.1640422736 |
- |
- |
PREDICTED: uncharacterized protein LOC105639921 [Jatropha curcas] |
| 17 |
Hb_000427_030 |
0.1649994621 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002835_410 |
0.1652982759 |
- |
- |
clathrin coat assembly protein ap-1, putative [Ricinus communis] |
| 19 |
Hb_004976_030 |
0.1653947833 |
- |
- |
hypothetical protein POPTR_0005s04700g [Populus trichocarpa] |
| 20 |
Hb_000115_330 |
0.1662822828 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |