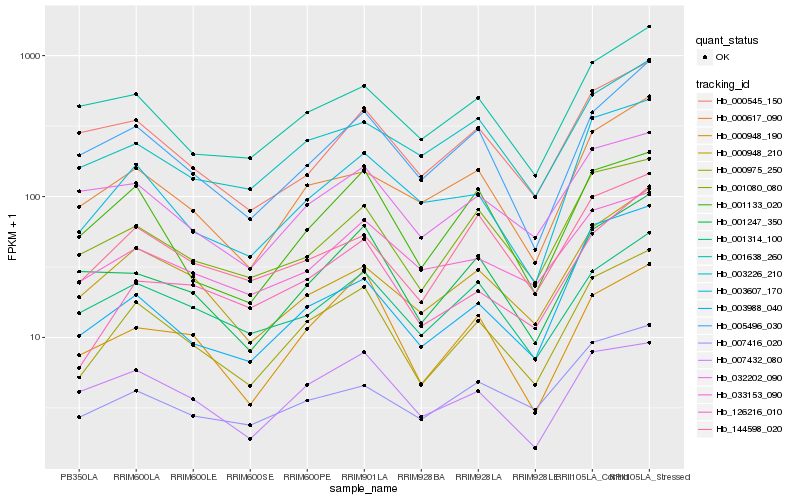
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_250 |
0.0 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 4, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_001080_080 |
0.1085643781 |
- |
- |
small nuclear ribonucleoprotein, putative [Ricinus communis] |
| 3 |
Hb_003607_170 |
0.1110195479 |
- |
- |
U6 snRNA-associated Sm-like protein LSm5 [Hevea brasiliensis] |
| 4 |
Hb_126216_010 |
0.1122986894 |
- |
- |
PREDICTED: 60S ribosomal protein L9 [Jatropha curcas] |
| 5 |
Hb_000948_190 |
0.1153676838 |
transcription factor |
TF Family: GNAT |
Glucosamine 6-phosphate N-acetyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000617_090 |
0.1157519372 |
- |
- |
hypoia-responsive family protein 4 [Hevea brasiliensis] |
| 7 |
Hb_144598_020 |
0.1157847077 |
- |
- |
PREDICTED: triphosphate tunel metalloenzyme 3 [Jatropha curcas] |
| 8 |
Hb_005496_030 |
0.1161660578 |
- |
- |
PREDICTED: 40S ribosomal protein S29-like [Malus domestica] |
| 9 |
Hb_001133_020 |
0.1161820501 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase complex SCF subunit sconB [Jatropha curcas] |
| 10 |
Hb_000948_210 |
0.1167976746 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001247_350 |
0.1172775263 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 11 [Jatropha curcas] |
| 12 |
Hb_001314_100 |
0.1224953029 |
- |
- |
hypothetical protein POPTR_0001s01820g [Populus trichocarpa] |
| 13 |
Hb_007432_080 |
0.1246149998 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1-like [Jatropha curcas] |
| 14 |
Hb_001638_260 |
0.1249069502 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1 [Vitis vinifera] |
| 15 |
Hb_003226_210 |
0.1271879426 |
- |
- |
hypothetical protein PRUPE_ppa013414mg [Prunus persica] |
| 16 |
Hb_007416_020 |
0.1274607411 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Cucumis sativus] |
| 17 |
Hb_033153_090 |
0.127912813 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 18 |
Hb_032202_090 |
0.1283235065 |
- |
- |
PREDICTED: ubiquitin-related modifier 1 homolog 2 [Jatropha curcas] |
| 19 |
Hb_003988_040 |
0.128449445 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 20 |
Hb_000545_150 |
0.131584805 |
- |
- |
PREDICTED: transcription factor BTF3 homolog 4-like [Malus domestica] |