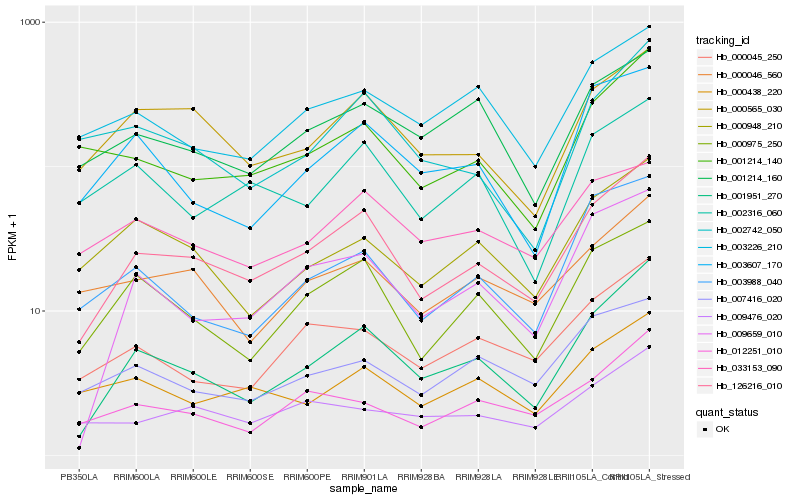
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126216_010 |
0.0 |
- |
- |
PREDICTED: 60S ribosomal protein L9 [Jatropha curcas] |
| 2 |
Hb_001951_270 |
0.0912768494 |
- |
- |
- |
| 3 |
Hb_000975_250 |
0.1122986894 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein 4, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_009659_010 |
0.1238094351 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 9 [Vitis vinifera] |
| 5 |
Hb_000565_030 |
0.1253486845 |
- |
- |
PREDICTED: 40S ribosomal protein S26-3-like [Jatropha curcas] |
| 6 |
Hb_000045_250 |
0.1326992613 |
- |
- |
PREDICTED: uncharacterized protein LOC105646572 isoform X1 [Jatropha curcas] |
| 7 |
Hb_012251_010 |
0.1372348167 |
- |
- |
- |
| 8 |
Hb_001214_140 |
0.1415753666 |
- |
- |
60S ribosomal protein L27a, putative [Ricinus communis] |
| 9 |
Hb_000046_560 |
0.1415923662 |
- |
- |
PREDICTED: uncharacterized protein LOC105118519 [Populus euphratica] |
| 10 |
Hb_003988_040 |
0.1431764439 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 11 |
Hb_000438_220 |
0.1447237759 |
- |
- |
PREDICTED: uncharacterized protein LOC105649213 [Jatropha curcas] |
| 12 |
Hb_009476_020 |
0.1452837315 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 13 |
Hb_003607_170 |
0.1459915923 |
- |
- |
U6 snRNA-associated Sm-like protein LSm5 [Hevea brasiliensis] |
| 14 |
Hb_007416_020 |
0.1461016886 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Cucumis sativus] |
| 15 |
Hb_003226_210 |
0.1466080023 |
- |
- |
hypothetical protein PRUPE_ppa013414mg [Prunus persica] |
| 16 |
Hb_002316_060 |
0.147110715 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 17 |
Hb_000948_210 |
0.1478595798 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001214_160 |
0.1488439889 |
- |
- |
60S ribosomal protein L27B [Hevea brasiliensis] |
| 19 |
Hb_033153_090 |
0.1495939935 |
- |
- |
PREDICTED: 28 kDa heat- and acid-stable phosphoprotein [Jatropha curcas] |
| 20 |
Hb_002742_050 |
0.1501849663 |
- |
- |
hypothetical protein CICLE_v10002863mg [Citrus clementina] |