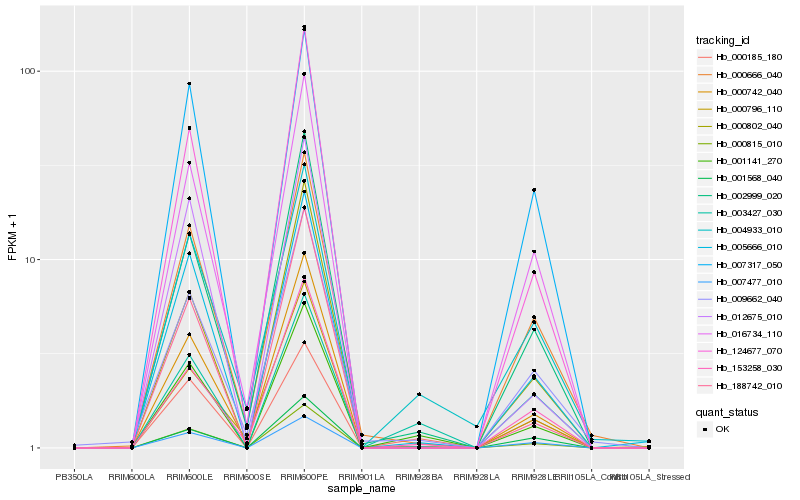
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000815_010 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105117338 [Populus euphratica] |
| 2 |
Hb_001141_270 |
0.0472375798 |
- |
- |
PREDICTED: uncharacterized protein LOC105632005 [Jatropha curcas] |
| 3 |
Hb_007477_010 |
0.0715164479 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA9 [Phoenix dactylifera] |
| 4 |
Hb_005666_010 |
0.074255521 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 5 |
Hb_002999_020 |
0.0856330442 |
- |
- |
PREDICTED: ribonuclease 3-like [Jatropha curcas] |
| 6 |
Hb_000742_040 |
0.0859891866 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 7 |
Hb_016734_110 |
0.0951967627 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 8 |
Hb_124677_070 |
0.100115938 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 9 |
Hb_000185_180 |
0.1011474102 |
- |
- |
hypothetical protein CISIN_1g037968mg, partial [Citrus sinensis] |
| 10 |
Hb_153258_030 |
0.1017193326 |
- |
- |
hypothetical protein RCOM_0853150 [Ricinus communis] |
| 11 |
Hb_000666_040 |
0.1025195314 |
- |
- |
PREDICTED: MLP-like protein 34 [Vitis vinifera] |
| 12 |
Hb_009662_040 |
0.1050948872 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 13 |
Hb_007317_050 |
0.1065294972 |
- |
- |
hypothetical protein POPTR_0009s16550g [Populus trichocarpa] |
| 14 |
Hb_000802_040 |
0.1137655542 |
- |
- |
PREDICTED: probable inositol transporter 2 [Jatropha curcas] |
| 15 |
Hb_001568_040 |
0.1163169993 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003427_030 |
0.1186520803 |
- |
- |
- |
| 17 |
Hb_004933_010 |
0.118726234 |
- |
- |
hypothetical protein POPTR_0018s10810g [Populus trichocarpa] |
| 18 |
Hb_000796_110 |
0.1213640504 |
- |
- |
PREDICTED: probable pectinesterase 53 [Populus euphratica] |
| 19 |
Hb_188742_010 |
0.124076517 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 20 |
Hb_012675_010 |
0.1246515421 |
- |
- |
zinc finger protein, putative [Ricinus communis] |