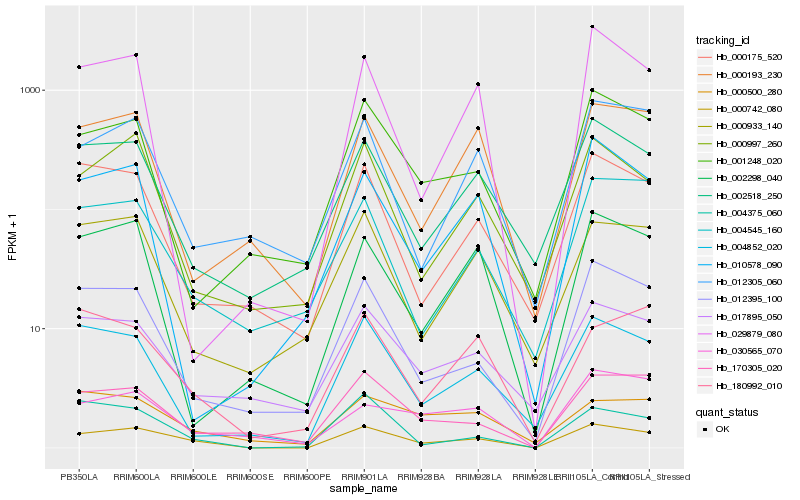
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_170305_020 |
0.1776629927 |
- |
- |
hypothetical protein JCGZ_23578 [Jatropha curcas] |
| 3 |
Hb_012395_100 |
0.1791884249 |
- |
- |
eukaryotic translation initiation factor 4e, putative [Ricinus communis] |
| 4 |
Hb_017895_050 |
0.1807833775 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: UPF0160 protein MYG1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_002518_250 |
0.1808174101 |
- |
- |
conserved hypothetical protein 10 [Hevea brasiliensis] |
| 6 |
Hb_004852_020 |
0.1842216309 |
- |
- |
- |
| 7 |
Hb_000997_260 |
0.1844415036 |
- |
- |
PREDICTED: rRNA-processing protein EFG1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004375_060 |
0.186163062 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000175_520 |
0.1877157353 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_001248_020 |
0.1895329599 |
- |
- |
PREDICTED: uncharacterized protein At2g23090-like [Jatropha curcas] |
| 11 |
Hb_000933_140 |
0.1911025877 |
- |
- |
PREDICTED: uncharacterized protein LOC105630501 [Jatropha curcas] |
| 12 |
Hb_000500_280 |
0.1931038466 |
- |
- |
Brassinosteroid-regulated protein BRU1 precursor, putative [Ricinus communis] |
| 13 |
Hb_180992_010 |
0.194229687 |
- |
- |
hypothetical protein POPTR_0010s24060g [Populus trichocarpa] |
| 14 |
Hb_000193_230 |
0.1951082457 |
- |
- |
PREDICTED: NEP1-interacting protein-like 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012305_060 |
0.1969262532 |
- |
- |
protein translocase, putative [Ricinus communis] |
| 16 |
Hb_004545_160 |
0.1977114808 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 17 |
Hb_002298_040 |
0.1981506989 |
- |
- |
PREDICTED: uncharacterized protein LOC105633642 [Jatropha curcas] |
| 18 |
Hb_029879_080 |
0.198560354 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_030565_070 |
0.2004733387 |
- |
- |
- |
| 20 |
Hb_010578_090 |
0.2006133434 |
- |
- |
HEV2.1 [Hevea brasiliensis] |