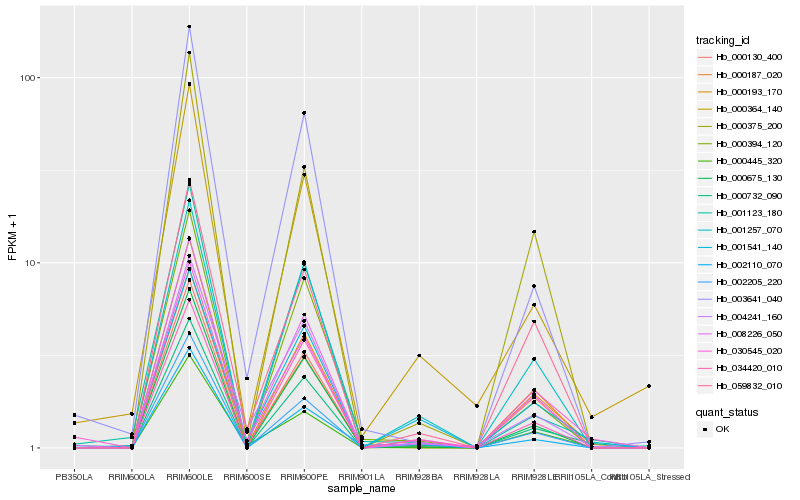
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_090 |
0.0 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 2 |
Hb_002205_220 |
0.088526829 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 3 |
Hb_000675_130 |
0.0888980303 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_008226_050 |
0.0927850701 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 5 |
Hb_000187_020 |
0.0998651398 |
- |
- |
Xanthine/uracil permease family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_000394_120 |
0.101358444 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2-like [Populus euphratica] |
| 7 |
Hb_000130_400 |
0.1014287487 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0001s38320g [Populus trichocarpa] |
| 8 |
Hb_001257_070 |
0.1015082548 |
- |
- |
PREDICTED: aspartic proteinase nepenthesin-1-like [Jatropha curcas] |
| 9 |
Hb_030545_020 |
0.1076715996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_034420_010 |
0.1078935974 |
- |
- |
glutathione S-transferase [Caragana korshinskii] |
| 11 |
Hb_000375_200 |
0.1147693683 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 12 |
Hb_000193_170 |
0.1158340124 |
transcription factor |
TF Family: MYB |
Myb domain 16-like protein [Theobroma cacao] |
| 13 |
Hb_001123_180 |
0.1160524178 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000445_320 |
0.1186054679 |
- |
- |
PREDICTED: wall-associated receptor kinase 2-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_004241_160 |
0.1186354608 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-7 [Jatropha curcas] |
| 16 |
Hb_001541_140 |
0.1192171548 |
- |
- |
PREDICTED: probable protein phosphatase 2C 14 [Jatropha curcas] |
| 17 |
Hb_002110_070 |
0.1219895379 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003641_040 |
0.1225492257 |
- |
- |
hypothetical protein POPTR_0006s14010g [Populus trichocarpa] |
| 19 |
Hb_059832_010 |
0.1243578934 |
- |
- |
PREDICTED: glutamate receptor 2.9-like [Jatropha curcas] |
| 20 |
Hb_000364_140 |
0.1249329323 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280-like [Jatropha curcas] |