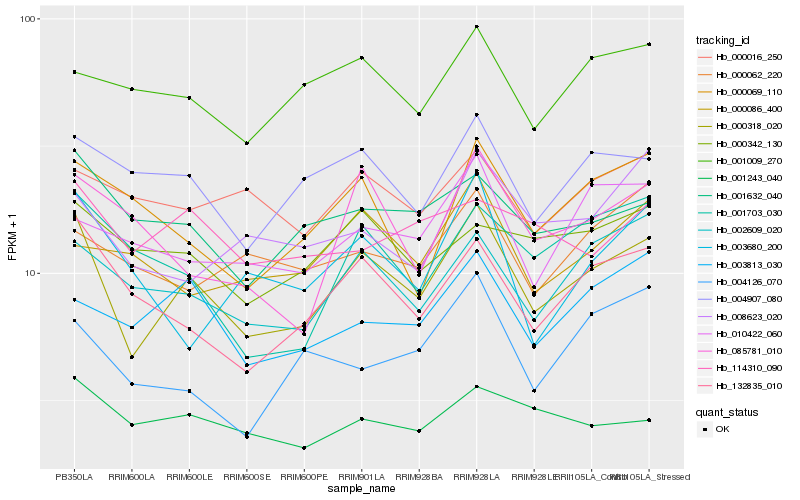
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_020 |
0.0 |
- |
- |
PREDICTED: sodium channel modifier 1-like [Jatropha curcas] |
| 2 |
Hb_132835_010 |
0.1073460922 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_002609_020 |
0.1131394214 |
- |
- |
PREDICTED: phosphatidylinositol N-acetylglucosaminyltransferase subunit P-like [Jatropha curcas] |
| 4 |
Hb_001632_040 |
0.1224532168 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_004907_080 |
0.1225163901 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_003813_030 |
0.1280910129 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 7 |
Hb_001703_030 |
0.1310427537 |
transcription factor |
TF Family: SET |
Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_001009_270 |
0.1321782136 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 6 homolog [Jatropha curcas] |
| 9 |
Hb_010422_060 |
0.1322694446 |
- |
- |
PREDICTED: GLABRA2 expression modulator [Jatropha curcas] |
| 10 |
Hb_000342_130 |
0.1327147775 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000069_110 |
0.133370327 |
- |
- |
PREDICTED: nephrocystin-3-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001243_040 |
0.1346791326 |
- |
- |
PREDICTED: uncharacterized protein LOC104104316 [Nicotiana tomentosiformis] |
| 13 |
Hb_085781_010 |
0.1363018573 |
- |
- |
PREDICTED: protein root UVB sensitive 4 [Jatropha curcas] |
| 14 |
Hb_008623_020 |
0.1365315013 |
- |
- |
PREDICTED: uncharacterized protein At3g17611 [Jatropha curcas] |
| 15 |
Hb_000016_250 |
0.1371419373 |
- |
- |
unnamed protein product [Malassezia sympodialis ATCC 42132] |
| 16 |
Hb_000086_400 |
0.1396428329 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 17 |
Hb_114310_090 |
0.1406847091 |
- |
- |
PREDICTED: low-temperature-induced cysteine proteinase [Jatropha curcas] |
| 18 |
Hb_003680_200 |
0.1408555662 |
- |
- |
PREDICTED: F-box protein PP2-A15 [Jatropha curcas] |
| 19 |
Hb_000062_220 |
0.1410945294 |
- |
- |
BnaC02g35870D [Brassica napus] |
| 20 |
Hb_004126_070 |
0.1412995751 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |