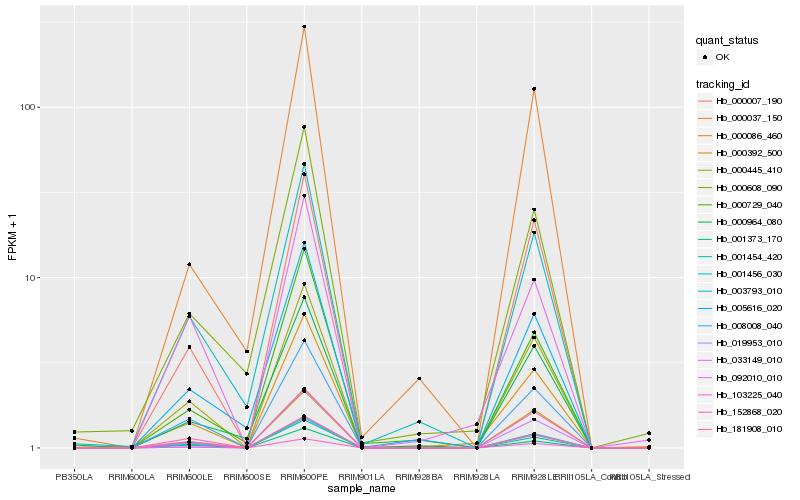
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_460 |
0.0 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 2 |
Hb_033149_010 |
0.0977924786 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_003793_010 |
0.0988540441 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 4 |
Hb_000445_410 |
0.09925955 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 5 |
Hb_000608_090 |
0.1032758153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_103225_040 |
0.1044292012 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 7 |
Hb_092010_010 |
0.10974606 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 8 |
Hb_001373_170 |
0.1113099576 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 9 |
Hb_152868_020 |
0.1139384951 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 10 |
Hb_008008_040 |
0.1143040326 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 11 |
Hb_000007_190 |
0.1159899426 |
- |
- |
PREDICTED: protein STRICTOSIDINE SYNTHASE-LIKE 10-like [Jatropha curcas] |
| 12 |
Hb_001456_030 |
0.1182174419 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 13 |
Hb_000037_150 |
0.1237685004 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 14 |
Hb_181908_010 |
0.1252232955 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 15 |
Hb_005616_020 |
0.1252298765 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_000964_080 |
0.1255022665 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 17 |
Hb_001454_420 |
0.1255221196 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 18 |
Hb_000729_040 |
0.1258342064 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 10 precursor [Populus trichocarpa] |
| 19 |
Hb_019953_010 |
0.1264320485 |
- |
- |
hypothetical protein CICLE_v10019877mg [Citrus clementina] |
| 20 |
Hb_000392_500 |
0.1276337369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |