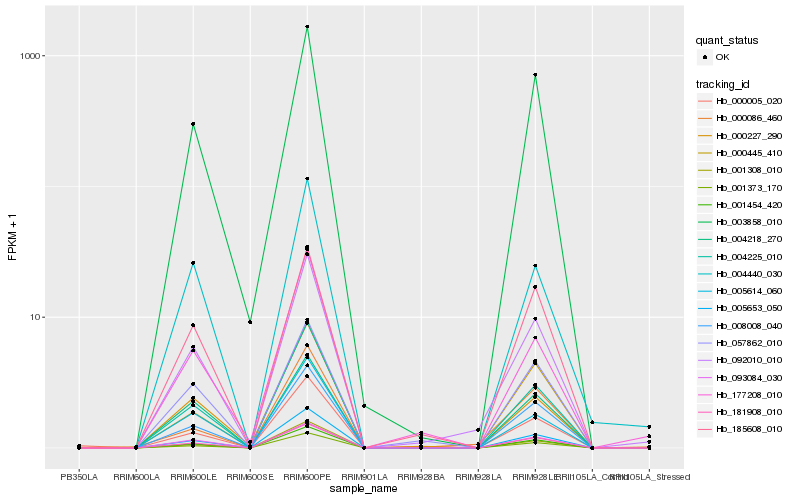
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092010_010 |
0.0 |
- |
- |
cysteine proteinase inhibitor 5-like [Cucumis sativus] |
| 2 |
Hb_001454_420 |
0.0853758514 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 3 |
Hb_005653_050 |
0.08816772 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-1,4-galactoside alpha-2,3-sialyltransferase [Jatropha curcas] |
| 4 |
Hb_004225_010 |
0.0896355678 |
- |
- |
- |
| 5 |
Hb_001373_170 |
0.0908204941 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 9 [Jatropha curcas] |
| 6 |
Hb_181908_010 |
0.09092321 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 7 |
Hb_008008_040 |
0.0957228375 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 8 |
Hb_000086_460 |
0.10974606 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.2-like [Jatropha curcas] |
| 9 |
Hb_001308_010 |
0.1101370587 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_093084_030 |
0.1103460697 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 11 |
Hb_003858_010 |
0.1105855213 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 12 |
Hb_005614_060 |
0.1111216291 |
- |
- |
Uncharacterized protein TCM_019750 [Theobroma cacao] |
| 13 |
Hb_057862_010 |
0.1114237391 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_177208_010 |
0.1118865126 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor, partial [Jatropha sp. CONN 198700232] |
| 15 |
Hb_004440_030 |
0.1121208884 |
transcription factor |
TF Family: MYB-related |
hypothetical protein POPTR_0005s12390g [Populus trichocarpa] |
| 16 |
Hb_185608_010 |
0.1163550227 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 17 |
Hb_000227_290 |
0.1163995087 |
- |
- |
PREDICTED: uncharacterized protein LOC105631957 [Jatropha curcas] |
| 18 |
Hb_000005_020 |
0.1191979444 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000445_410 |
0.1215704784 |
- |
- |
hypothetical protein EUGRSUZ_F02776 [Eucalyptus grandis] |
| 20 |
Hb_004218_270 |
0.1217300969 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |