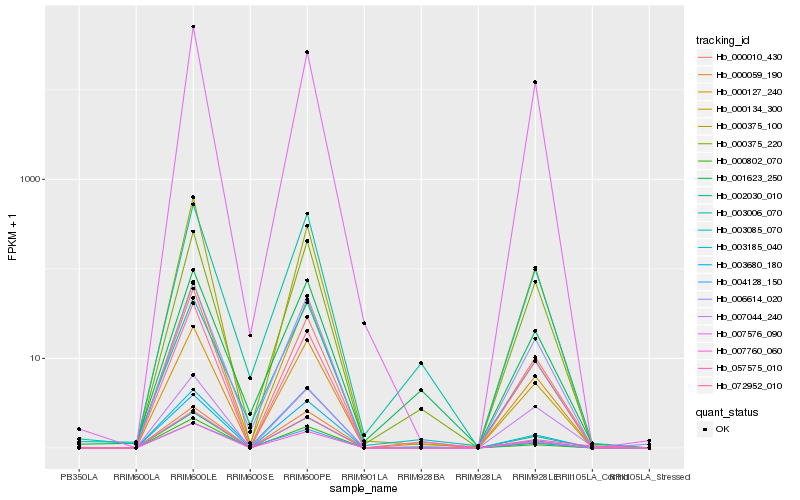
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_057575_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002030_010 |
0.1041938574 |
- |
- |
hypothetical protein JCGZ_18681 [Jatropha curcas] |
| 3 |
Hb_000059_190 |
0.1103331832 |
- |
- |
PREDICTED: uncharacterized protein LOC105121874 [Populus euphratica] |
| 4 |
Hb_007760_060 |
0.1129213388 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0010s17470g [Populus trichocarpa] |
| 5 |
Hb_000802_070 |
0.1133657675 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: eugenol synthase 1-like [Jatropha curcas] |
| 6 |
Hb_000127_240 |
0.1141085417 |
- |
- |
SAUR39 [Populus tomentosa] |
| 7 |
Hb_006614_020 |
0.1165513015 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 8 |
Hb_003006_070 |
0.1209546444 |
- |
- |
aquaporin [Manihot esculenta] |
| 9 |
Hb_000375_220 |
0.1251310698 |
- |
- |
RecName: Full=2-methylbutanal oxime monooxygenase; AltName: Full=Cytochrome P450 71E7 [Manihot esculenta] |
| 10 |
Hb_000134_300 |
0.1271845841 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_000010_430 |
0.1274626524 |
- |
- |
Esterase precursor, putative [Ricinus communis] |
| 12 |
Hb_003085_070 |
0.1302336512 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000375_100 |
0.1303761431 |
- |
- |
RecName: Full=Valine N-monooxygenase 1; AltName: Full=Cytochrome P450 79D1 [Manihot esculenta] |
| 14 |
Hb_003185_040 |
0.1351467954 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 15 |
Hb_007044_240 |
0.1359067383 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004128_150 |
0.1409379614 |
- |
- |
hypothetical protein POPTR_0016s08360g [Populus trichocarpa] |
| 17 |
Hb_007576_090 |
0.1416416442 |
- |
- |
lipid transfer protein precursor [Gossypium herbaceum subsp. africanum] |
| 18 |
Hb_001623_250 |
0.1459129984 |
- |
- |
PREDICTED: probable pectate lyase 8 [Jatropha curcas] |
| 19 |
Hb_072952_010 |
0.1459694833 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g18430-like [Populus euphratica] |
| 20 |
Hb_003680_180 |
0.1476946102 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |