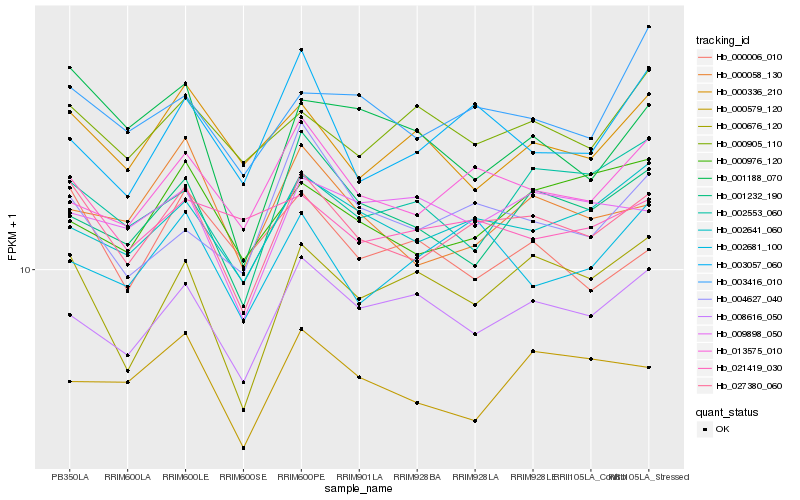
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_060 |
0.0 |
- |
- |
PREDICTED: putative ALA-interacting subunit 2 [Jatropha curcas] |
| 2 |
Hb_002641_060 |
0.0703494474 |
- |
- |
prefoldin subunit, putative [Ricinus communis] |
| 3 |
Hb_013575_010 |
0.070877353 |
- |
- |
Hepatocyte growth factor-regulated tyrosine kinase substrate, putative [Ricinus communis] |
| 4 |
Hb_008616_050 |
0.0732501315 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 5 |
Hb_000058_130 |
0.076814745 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 6 |
Hb_000676_120 |
0.0770065232 |
- |
- |
- |
| 7 |
Hb_001232_190 |
0.0785302272 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003416_010 |
0.0813761613 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 13 homolog B [Malus domestica] |
| 9 |
Hb_009898_050 |
0.0813882843 |
- |
- |
PREDICTED: beta-taxilin [Jatropha curcas] |
| 10 |
Hb_004627_040 |
0.0820884657 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 11 |
Hb_000579_120 |
0.0824546408 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 12 |
Hb_002681_100 |
0.0833367349 |
- |
- |
PREDICTED: glucuronokinase 1 [Jatropha curcas] |
| 13 |
Hb_021419_030 |
0.0842932661 |
- |
- |
hypothetical protein glysoja_023295 [Glycine soja] |
| 14 |
Hb_000905_110 |
0.0843063234 |
- |
- |
arginine/serine rich splicing factor sf4/14, putative [Ricinus communis] |
| 15 |
Hb_000336_210 |
0.0845582332 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000976_120 |
0.084731467 |
- |
- |
PREDICTED: DNA polymerase zeta processivity subunit [Jatropha curcas] |
| 17 |
Hb_003057_060 |
0.0850021659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002553_060 |
0.0864315742 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit beta, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001188_070 |
0.0864776015 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 20 |
Hb_000006_010 |
0.0875807091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |