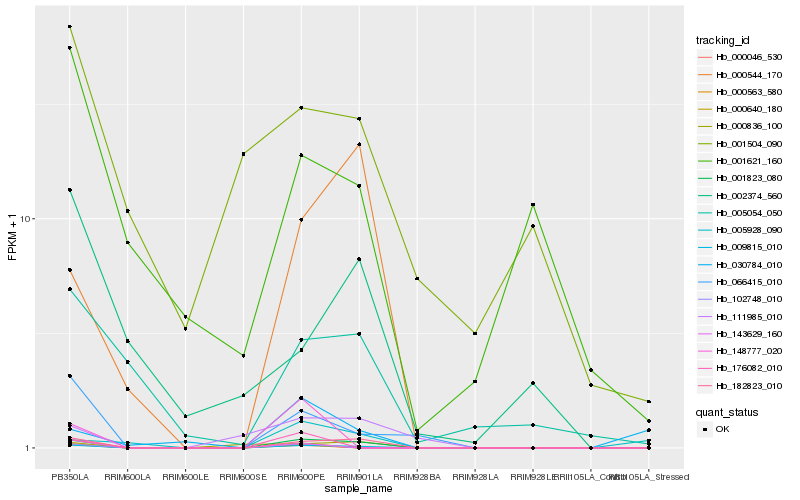
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009815_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001823_080 |
0.0583816029 |
- |
- |
hypothetical protein VITISV_012142 [Vitis vinifera] |
| 3 |
Hb_182823_010 |
0.1306784172 |
- |
- |
PREDICTED: thylakoidal processing peptidase 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_066415_010 |
0.2847649555 |
- |
- |
- |
| 5 |
Hb_000640_180 |
0.352321145 |
- |
- |
enzyme inhibitor, putative [Ricinus communis] |
| 6 |
Hb_000563_580 |
0.3523403938 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000046_530 |
0.3524506901 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF086 [Jatropha curcas] |
| 8 |
Hb_000544_170 |
0.3525819703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_102748_010 |
0.3544839916 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 10 |
Hb_005054_050 |
0.3598834382 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 11 |
Hb_111985_010 |
0.3613046506 |
- |
- |
- |
| 12 |
Hb_143629_160 |
0.3628050624 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 13 |
Hb_000836_100 |
0.3695179674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_176082_010 |
0.3849837707 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 15 |
Hb_001621_160 |
0.3915124922 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 16 |
Hb_005928_090 |
0.3991517871 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like isoform X3 [Jatropha curcas] |
| 17 |
Hb_148777_020 |
0.4003748926 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 18 |
Hb_030784_010 |
0.4065716509 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 19 |
Hb_001504_090 |
0.4075303511 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_002374_560 |
0.4092061943 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH11-like [Populus euphratica] |