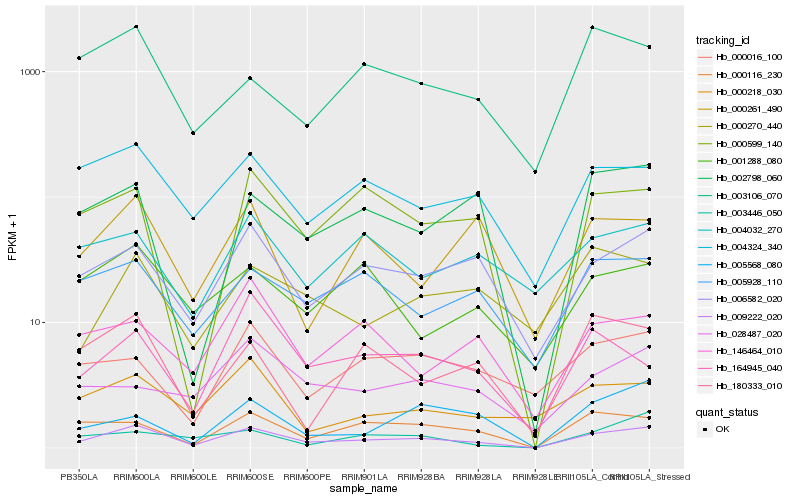
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009222_020 |
0.0 |
- |
- |
hypothetical protein AALP_AA5G087000 [Arabis alpina] |
| 2 |
Hb_006582_020 |
0.1684871318 |
- |
- |
Zinc finger protein ZFPM1 [Theobroma cacao] |
| 3 |
Hb_004324_340 |
0.183818981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000599_140 |
0.1871616599 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 4-like [Populus euphratica] |
| 5 |
Hb_000116_230 |
0.1894636144 |
- |
- |
- |
| 6 |
Hb_002798_060 |
0.1949563147 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 1-like [Vitis vinifera] |
| 7 |
Hb_005928_110 |
0.1969142136 |
- |
- |
PREDICTED: PRA1 family protein F2-like [Jatropha curcas] |
| 8 |
Hb_000218_030 |
0.2083200449 |
- |
- |
- |
| 9 |
Hb_000270_440 |
0.2099339291 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0015s08130g [Populus trichocarpa] |
| 10 |
Hb_180333_010 |
0.2100209552 |
- |
- |
- |
| 11 |
Hb_001288_080 |
0.2118880909 |
- |
- |
PREDICTED: probable protein phosphatase 2C 50 [Populus euphratica] |
| 12 |
Hb_000261_490 |
0.2118992993 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 13 |
Hb_005568_080 |
0.2125176304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000016_100 |
0.2128734349 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 15 |
Hb_164945_040 |
0.2137069143 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 16 |
Hb_003106_070 |
0.2139328747 |
- |
- |
PREDICTED: uncharacterized protein LOC105167687 [Sesamum indicum] |
| 17 |
Hb_146464_010 |
0.2146854613 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_003446_050 |
0.2147892633 |
- |
- |
hypothetical protein B456_002G034000, partial [Gossypium raimondii] |
| 19 |
Hb_028487_020 |
0.2154253665 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 20 |
Hb_004032_270 |
0.2166765825 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |