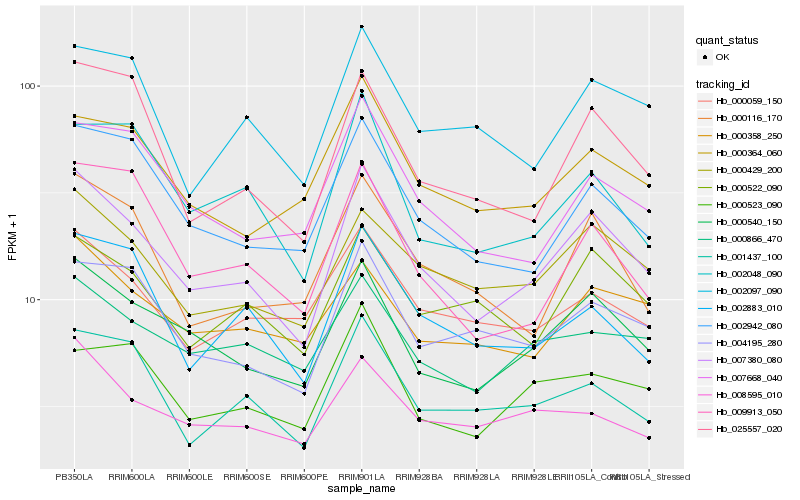
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007380_080 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: myb family transcription factor APL-like [Jatropha curcas] |
| 2 |
Hb_000540_150 |
0.0754812901 |
- |
- |
PREDICTED: uncharacterized protein YKR070W [Jatropha curcas] |
| 3 |
Hb_000429_200 |
0.0941779873 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 4 |
Hb_002942_080 |
0.1002396566 |
- |
- |
PREDICTED: signal recognition particle subunit SRP68 [Jatropha curcas] |
| 5 |
Hb_025557_020 |
0.1005130887 |
- |
- |
PREDICTED: activating signal cointegrator 1 [Jatropha curcas] |
| 6 |
Hb_004195_280 |
0.1040470218 |
- |
- |
PREDICTED: CDK5RAP1-like protein [Jatropha curcas] |
| 7 |
Hb_000116_170 |
0.106287901 |
- |
- |
hypothetical protein JCGZ_22392 [Jatropha curcas] |
| 8 |
Hb_007668_040 |
0.1063724144 |
- |
- |
PREDICTED: uncharacterized protein LOC103409082 [Malus domestica] |
| 9 |
Hb_001437_100 |
0.1063910124 |
- |
- |
PREDICTED: DNA-binding protein REB1-like [Jatropha curcas] |
| 10 |
Hb_000522_090 |
0.1075593964 |
- |
- |
PREDICTED: actin-related protein 9 isoform X1 [Nicotiana tomentosiformis] |
| 11 |
Hb_000364_060 |
0.1103811147 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 12 |
Hb_002097_090 |
0.1114826543 |
- |
- |
Splicing factor 3A subunit, putative [Ricinus communis] |
| 13 |
Hb_000866_470 |
0.11206254 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 14 |
Hb_002048_090 |
0.1124710977 |
- |
- |
PREDICTED: uncharacterized protein LOC105644513 [Jatropha curcas] |
| 15 |
Hb_002883_010 |
0.112895966 |
- |
- |
PREDICTED: putative methyltransferase NSUN6 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000059_150 |
0.1138888114 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_009913_050 |
0.1158688903 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 18 |
Hb_008595_010 |
0.1165024489 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06710, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000358_250 |
0.1166198148 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 20 |
Hb_000523_090 |
0.1175361175 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |