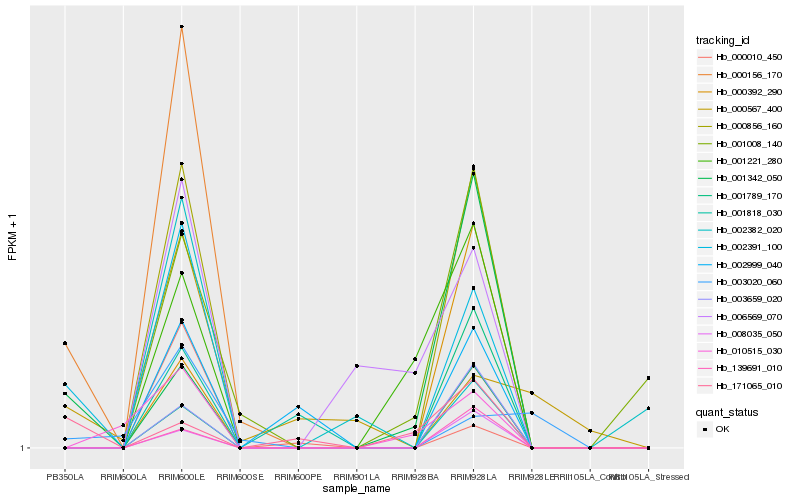
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002391_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0007s09360g [Populus trichocarpa] |
| 2 |
Hb_000856_160 |
0.2700096403 |
- |
- |
- |
| 3 |
Hb_000156_170 |
0.2827883738 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Prunus mume] |
| 4 |
Hb_003659_020 |
0.3418918921 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 5 |
Hb_008035_050 |
0.3450786409 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 6 |
Hb_139691_010 |
0.3513597043 |
- |
- |
- |
| 7 |
Hb_001342_050 |
0.3567309807 |
- |
- |
- |
| 8 |
Hb_002999_040 |
0.366580475 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001818_030 |
0.3726499061 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 10 |
Hb_000010_450 |
0.3729751293 |
- |
- |
purine permease, putative [Ricinus communis] |
| 11 |
Hb_000392_290 |
0.3845980666 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_171065_010 |
0.4037322537 |
- |
- |
Ribonuclease H protein [Theobroma cacao] |
| 13 |
Hb_010515_030 |
0.4061489552 |
- |
- |
PREDICTED: G2/mitotic-specific cyclin S13-7 [Jatropha curcas] |
| 14 |
Hb_001221_280 |
0.4064645431 |
- |
- |
- |
| 15 |
Hb_001789_170 |
0.4155826978 |
- |
- |
hypothetical protein JCGZ_08269 [Jatropha curcas] |
| 16 |
Hb_000567_400 |
0.4159486427 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 17 |
Hb_001008_140 |
0.419665931 |
- |
- |
- |
| 18 |
Hb_006569_070 |
0.4208849266 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_003020_060 |
0.4209289549 |
- |
- |
PREDICTED: low-temperature-induced 65 kDa protein [Jatropha curcas] |
| 20 |
Hb_002382_020 |
0.4223005755 |
- |
- |
PREDICTED: 3beta-hydroxysteroid-dehydrogenase/decarboxylase isoform 2 [Jatropha curcas] |