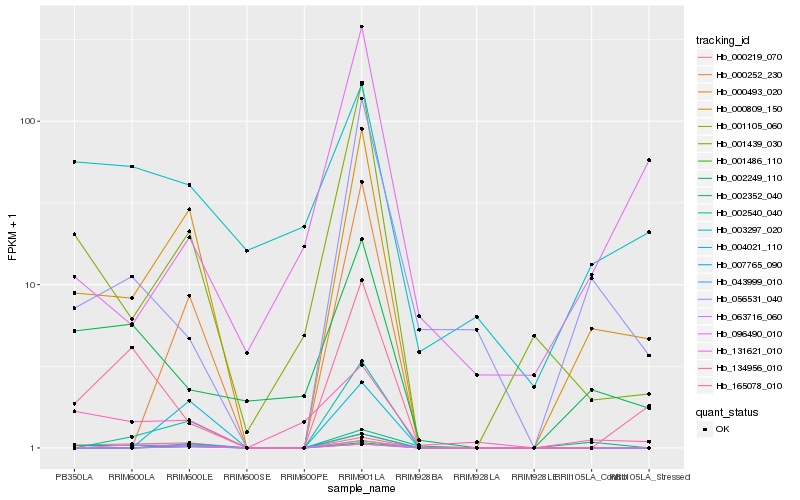
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000493_020 |
0.0 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_001439_030 |
0.2401182683 |
- |
- |
- |
| 3 |
Hb_002540_040 |
0.2555456162 |
- |
- |
BnaC05g39550D [Brassica napus] |
| 4 |
Hb_000219_070 |
0.266529559 |
- |
- |
- |
| 5 |
Hb_131621_010 |
0.2970248181 |
- |
- |
hypothetical protein MIMGU_mgv1a004274mg [Erythranthe guttata] |
| 6 |
Hb_007765_090 |
0.2973515436 |
- |
- |
unknown [Glycine max] |
| 7 |
Hb_043999_010 |
0.2974525131 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 8 |
Hb_001486_110 |
0.3017945069 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-11-like [Jatropha curcas] |
| 9 |
Hb_001105_060 |
0.3028272326 |
- |
- |
hypothetical protein CISIN_1g0045352mg, partial [Citrus sinensis] |
| 10 |
Hb_000809_150 |
0.303293619 |
- |
- |
- |
| 11 |
Hb_056531_040 |
0.3081700669 |
- |
- |
PREDICTED: uncharacterized protein LOC103494239 [Cucumis melo] |
| 12 |
Hb_000252_230 |
0.3096168631 |
- |
- |
hypothetical protein B456_008G160800 [Gossypium raimondii] |
| 13 |
Hb_134956_010 |
0.3177632751 |
- |
- |
PREDICTED: (S)-coclaurine N-methyltransferase [Jatropha curcas] |
| 14 |
Hb_004021_110 |
0.3226722397 |
- |
- |
- |
| 15 |
Hb_063716_060 |
0.3239897439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002249_110 |
0.3245516481 |
- |
- |
PREDICTED: centromere protein S isoform X1 [Jatropha curcas] |
| 17 |
Hb_002352_040 |
0.3297975818 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 18 |
Hb_165078_010 |
0.3300073143 |
- |
- |
- |
| 19 |
Hb_003297_020 |
0.3393140766 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 20 |
Hb_096490_010 |
0.3396201305 |
- |
- |
hypothetical protein CICLE_v100210772mg, partial [Citrus clementina] |