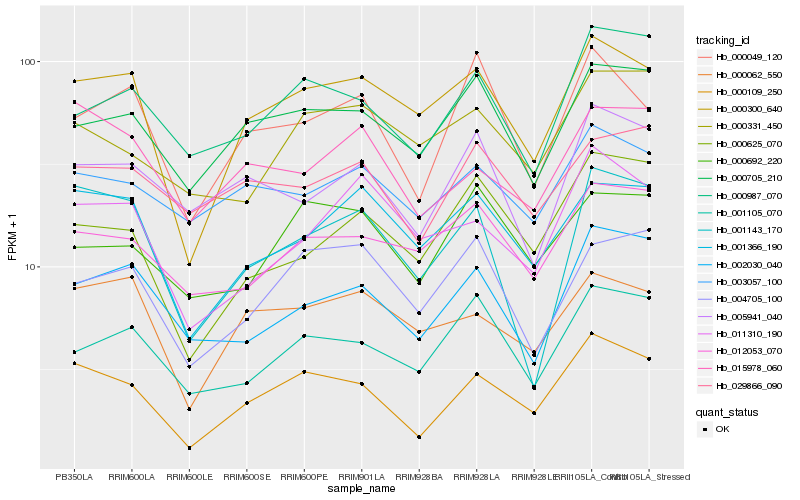
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000109_250 |
0.0 |
- |
- |
- |
| 2 |
Hb_000300_640 |
0.1124827548 |
- |
- |
PREDICTED: uncharacterized protein LOC100855285 [Vitis vinifera] |
| 3 |
Hb_000987_070 |
0.1234065415 |
- |
- |
Inflorescence meristem receptor-like kinase 2 isoform 1 [Theobroma cacao] |
| 4 |
Hb_000705_210 |
0.1275173948 |
- |
- |
PREDICTED: ras-related protein Rab2BV [Jatropha curcas] |
| 5 |
Hb_015978_060 |
0.1294521631 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_002030_040 |
0.1313648483 |
- |
- |
hypothetical protein JCGZ_18677 [Jatropha curcas] |
| 7 |
Hb_001366_190 |
0.1339604869 |
- |
- |
PREDICTED: uncharacterized protein LOC105632829 [Jatropha curcas] |
| 8 |
Hb_000062_550 |
0.1356869498 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001143_170 |
0.1362451092 |
- |
- |
nucellin, putative [Ricinus communis] |
| 10 |
Hb_001105_070 |
0.1367306498 |
- |
- |
PREDICTED: uncharacterized protein LOC105632829 [Jatropha curcas] |
| 11 |
Hb_000049_120 |
0.1367948183 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 12 |
Hb_003057_100 |
0.1369727725 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: S-adenosylmethionine mitochondrial carrier protein-like [Malus domestica] |
| 13 |
Hb_012053_070 |
0.1369817754 |
- |
- |
PREDICTED: uncharacterized protein LOC105637344 [Jatropha curcas] |
| 14 |
Hb_000625_070 |
0.1390670789 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_029866_090 |
0.1419832219 |
- |
- |
PREDICTED: uncharacterized protein LOC105648377 [Jatropha curcas] |
| 16 |
Hb_000331_450 |
0.1431347189 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 6 homolog [Jatropha curcas] |
| 17 |
Hb_011310_190 |
0.1434162289 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000692_220 |
0.143612663 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 19 |
Hb_005941_040 |
0.1439177736 |
- |
- |
PREDICTED: CASP-like protein 5A2 [Jatropha curcas] |
| 20 |
Hb_004705_100 |
0.1446935905 |
- |
- |
PREDICTED: inactive leucine-rich repeat receptor-like protein kinase CORYNE [Jatropha curcas] |