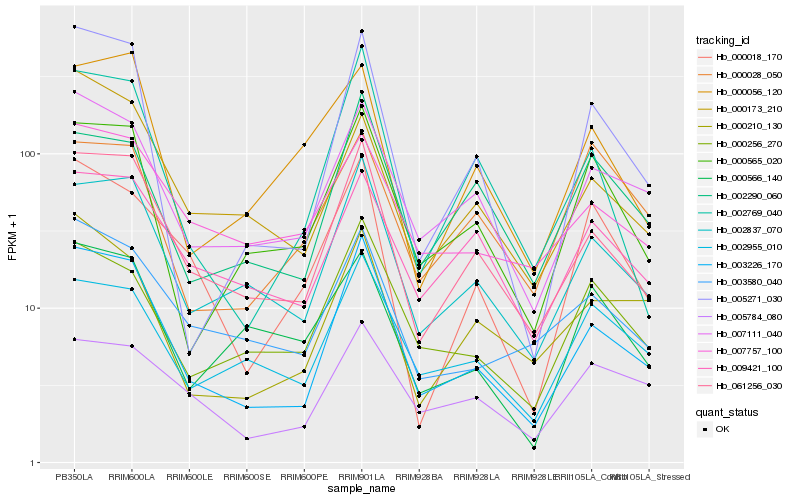
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000018_170 |
0.0 |
- |
- |
PREDICTED: bark storage protein A-like [Jatropha curcas] |
| 2 |
Hb_061256_030 |
0.1565565179 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 3 |
Hb_003226_170 |
0.1616718598 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_007111_040 |
0.1762532979 |
- |
- |
PREDICTED: uncharacterized protein LOC105628142 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000256_270 |
0.1819146414 |
- |
- |
PREDICTED: large proline-rich protein bag6-B isoform X1 [Jatropha curcas] |
| 6 |
Hb_003580_040 |
0.182259246 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2-like [Jatropha curcas] |
| 7 |
Hb_005784_080 |
0.1836043928 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_002837_070 |
0.1848939154 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 8-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_009421_100 |
0.1899651496 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 10 |
Hb_000566_140 |
0.1910635656 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 16 [Jatropha curcas] |
| 11 |
Hb_002769_040 |
0.191212788 |
- |
- |
farnesyl-diphosphate farnesyltransferase, putative [Ricinus communis] |
| 12 |
Hb_000565_020 |
0.1931475009 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 13 |
Hb_002955_010 |
0.1967302182 |
- |
- |
PREDICTED: Werner syndrome ATP-dependent helicase homolog [Jatropha curcas] |
| 14 |
Hb_000028_050 |
0.19956378 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 15 |
Hb_002290_060 |
0.2006725677 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 16 |
Hb_000056_120 |
0.2010045342 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 17 |
Hb_000210_130 |
0.2015268368 |
- |
- |
PREDICTED: chaperone protein ClpB4, mitochondrial [Vitis vinifera] |
| 18 |
Hb_007757_100 |
0.202123488 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 19 |
Hb_000173_210 |
0.2030706033 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 20 |
Hb_005271_030 |
0.2042241538 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |