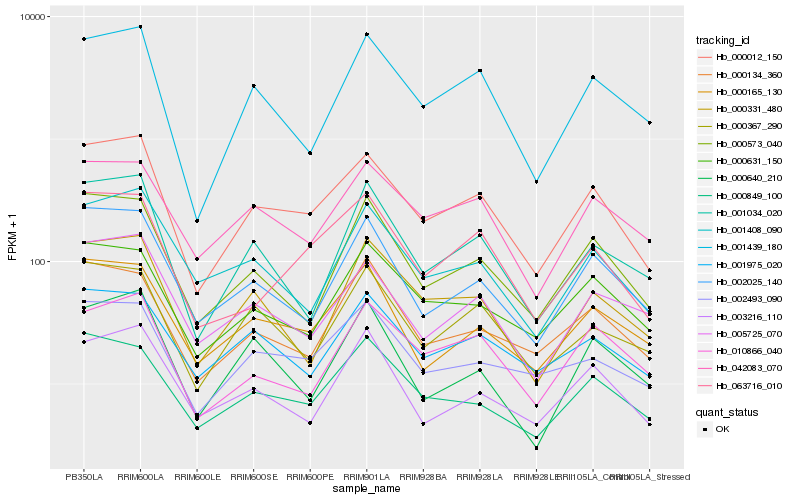
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_150 |
0.0 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 2 |
Hb_001439_180 |
0.10671588 |
- |
- |
hypothetical protein POPTR_0004s13090g [Populus trichocarpa] |
| 3 |
Hb_002025_140 |
0.1076647368 |
- |
- |
PREDICTED: UBX domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_042083_070 |
0.1096089455 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 5 |
Hb_063716_010 |
0.1114722695 |
- |
- |
phopholipase d alpha, putative [Ricinus communis] |
| 6 |
Hb_000573_040 |
0.1119411961 |
- |
- |
hypothetical protein JCGZ_19524 [Jatropha curcas] |
| 7 |
Hb_005725_070 |
0.112727014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000165_130 |
0.112990803 |
- |
- |
PREDICTED: uncharacterized protein At5g39865-like [Jatropha curcas] |
| 9 |
Hb_001408_090 |
0.1145994218 |
- |
- |
PREDICTED: uncharacterized protein LOC105629659 [Jatropha curcas] |
| 10 |
Hb_000331_480 |
0.1152774092 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3 [Jatropha curcas] |
| 11 |
Hb_002493_090 |
0.1170261287 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000367_290 |
0.1179823657 |
- |
- |
PREDICTED: uncharacterized protein LOC105640583 [Jatropha curcas] |
| 13 |
Hb_001034_020 |
0.11943128 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 14 |
Hb_000631_150 |
0.1196208495 |
- |
- |
PREDICTED: RNA polymerase-associated protein LEO1 [Jatropha curcas] |
| 15 |
Hb_000849_100 |
0.1197728528 |
- |
- |
PREDICTED: uncharacterized protein LOC105644134 [Jatropha curcas] |
| 16 |
Hb_010866_040 |
0.1220441145 |
- |
- |
PREDICTED: metal transporter Nramp2-like [Jatropha curcas] |
| 17 |
Hb_003216_110 |
0.1225433873 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 18 |
Hb_001975_020 |
0.1254098846 |
- |
- |
PREDICTED: uncharacterized protein LOC105637186 [Jatropha curcas] |
| 19 |
Hb_000640_210 |
0.1267290656 |
- |
- |
hypothetical protein JCGZ_14045 [Jatropha curcas] |
| 20 |
Hb_000134_360 |
0.1269090924 |
- |
- |
PREDICTED: protein gar2 [Jatropha curcas] |