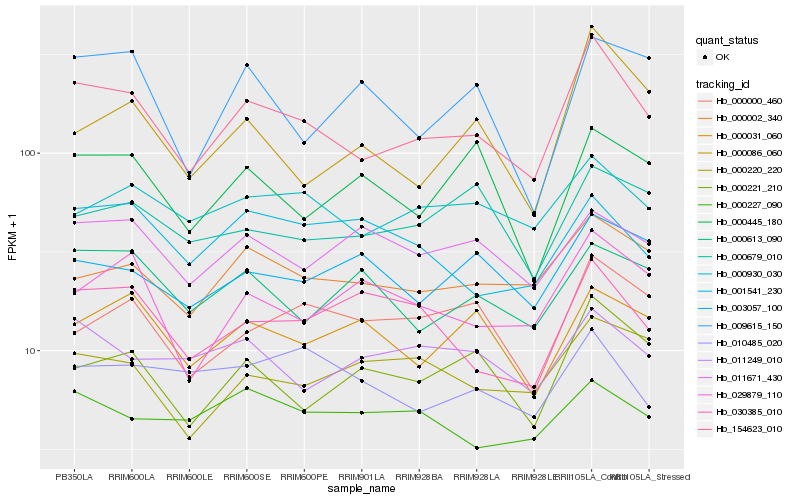
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_154623_010 |
0.0 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000002_340 |
0.1173161403 |
- |
- |
PREDICTED: peroxisomal bifunctional enzyme-like [Jatropha curcas] |
| 3 |
Hb_000221_210 |
0.1216479186 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 4 |
Hb_000930_030 |
0.1271365782 |
- |
- |
PREDICTED: uncharacterized protein LOC105629651 [Jatropha curcas] |
| 5 |
Hb_029879_110 |
0.1303966644 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g61870, mitochondrial [Jatropha curcas] |
| 6 |
Hb_030385_010 |
0.1345637673 |
- |
- |
PREDICTED: zinc-binding alcohol dehydrogenase domain-containing protein 2-like [Jatropha curcas] |
| 7 |
Hb_000000_460 |
0.1346921898 |
- |
- |
PREDICTED: peroxisomal membrane protein 11A [Jatropha curcas] |
| 8 |
Hb_000220_220 |
0.1355674598 |
- |
- |
PREDICTED: uncharacterized protein LOC105636907 [Jatropha curcas] |
| 9 |
Hb_000086_060 |
0.1359856734 |
- |
- |
PREDICTED: TOM1-like protein 2 isoform X2 [Populus euphratica] |
| 10 |
Hb_000613_090 |
0.1366859555 |
- |
- |
PREDICTED: uncharacterized protein LOC105641542 isoform X1 [Jatropha curcas] |
| 11 |
Hb_011249_010 |
0.1367578079 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 35, NatC auxiliary subunit isoform X1 [Jatropha curcas] |
| 12 |
Hb_000031_060 |
0.1383837263 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF12-A-like [Jatropha curcas] |
| 13 |
Hb_003057_100 |
0.1393748535 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: S-adenosylmethionine mitochondrial carrier protein-like [Malus domestica] |
| 14 |
Hb_011671_430 |
0.1402036962 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 15 |
Hb_001541_230 |
0.1408377875 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000679_010 |
0.1411233838 |
- |
- |
PREDICTED: uncharacterized protein LOC105637663 [Jatropha curcas] |
| 17 |
Hb_000445_180 |
0.1427663455 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 18 |
Hb_010485_020 |
0.143798165 |
- |
- |
PREDICTED: thaumatin-like protein 1b isoform X1 [Jatropha curcas] |
| 19 |
Hb_000227_090 |
0.1438379923 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 catalytic subunit [Jatropha curcas] |
| 20 |
Hb_009615_150 |
0.1451147761 |
- |
- |
PREDICTED: transcription elongation factor SPT4 homolog 2 [Jatropha curcas] |