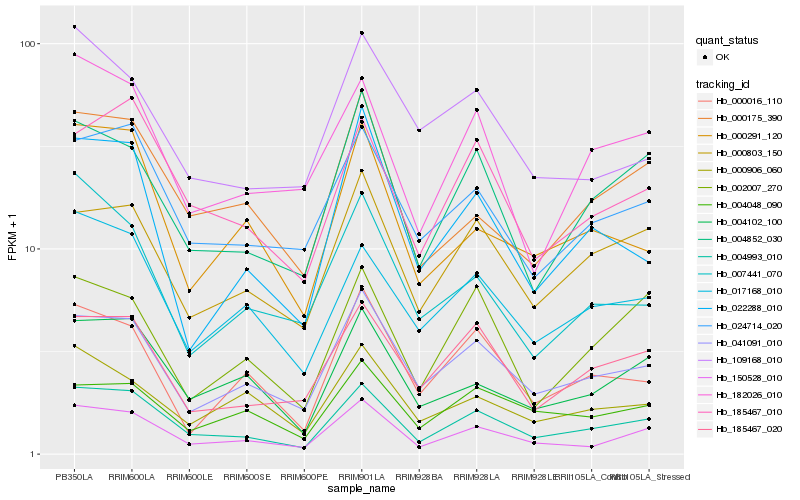
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150528_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004993_010 |
0.0900212274 |
- |
- |
PREDICTED: uncharacterized protein LOC104889591 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_004102_100 |
0.1130128266 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 4 |
Hb_041091_010 |
0.1131073306 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 5 |
Hb_004852_030 |
0.1239860204 |
- |
- |
PREDICTED: uncharacterized protein LOC105639923 [Jatropha curcas] |
| 6 |
Hb_000175_390 |
0.1267819902 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 7 |
Hb_024714_020 |
0.1370467145 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 8 |
Hb_000906_060 |
0.1377275065 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g08820 [Populus euphratica] |
| 9 |
Hb_000291_120 |
0.1388923137 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 17 [Jatropha curcas] |
| 10 |
Hb_007441_070 |
0.1426574456 |
- |
- |
hypothetical protein PRUPE_ppa004494mg [Prunus persica] |
| 11 |
Hb_000803_150 |
0.1434393051 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X2 [Jatropha curcas] |
| 12 |
Hb_017168_010 |
0.1445738362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_185467_020 |
0.144581997 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 14 |
Hb_185467_010 |
0.1445843597 |
- |
- |
- |
| 15 |
Hb_109168_010 |
0.1448086978 |
- |
- |
protein farnesyltransferase alpha subunit, putative [Ricinus communis] |
| 16 |
Hb_004048_090 |
0.1448242184 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g53330 [Jatropha curcas] |
| 17 |
Hb_002007_270 |
0.1450541811 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein At3g05340-like [Jatropha curcas] |
| 18 |
Hb_182026_010 |
0.1465652965 |
- |
- |
hypothetical protein CICLE_v10003487mg, partial [Citrus clementina] |
| 19 |
Hb_000016_110 |
0.1468226787 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 20 |
Hb_022288_010 |
0.1470506857 |
- |
- |
ubiquitin ligase E3 alpha, putative [Ricinus communis] |