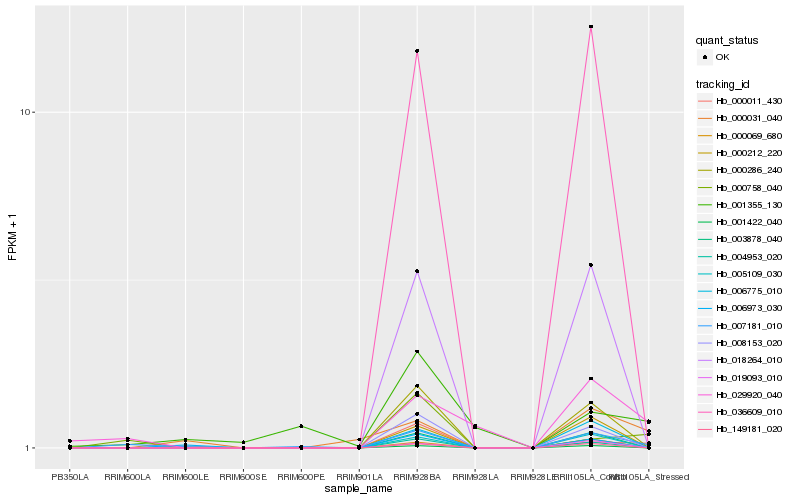
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007181_010 |
0.0 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 2 |
Hb_000011_430 |
0.3377437673 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_006973_030 |
0.3390998412 |
- |
- |
- |
| 4 |
Hb_003878_040 |
0.3410683711 |
- |
- |
- |
| 5 |
Hb_000286_240 |
0.341610086 |
- |
- |
- |
| 6 |
Hb_149181_020 |
0.3417967964 |
- |
- |
- |
| 7 |
Hb_018264_010 |
0.341862029 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 8 |
Hb_001422_040 |
0.3419300178 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 9 |
Hb_004953_020 |
0.3437899341 |
- |
- |
PREDICTED: uncharacterized protein LOC104234330 [Nicotiana sylvestris] |
| 10 |
Hb_008153_020 |
0.345411118 |
- |
- |
- |
| 11 |
Hb_036609_010 |
0.3456018756 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 12 |
Hb_029920_040 |
0.3472873339 |
- |
- |
PREDICTED: uncharacterized protein LOC105635837 [Jatropha curcas] |
| 13 |
Hb_005109_030 |
0.3480116048 |
- |
- |
PREDICTED: metal tolerance protein C2 [Jatropha curcas] |
| 14 |
Hb_000758_040 |
0.3499453989 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 15 |
Hb_001355_130 |
0.3525582903 |
- |
- |
PREDICTED: uncharacterized protein LOC105137896 [Populus euphratica] |
| 16 |
Hb_000212_220 |
0.3552599217 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 17 |
Hb_006775_010 |
0.3562903991 |
transcription factor |
TF Family: M-type |
AGAMOUS [Populus yunnanensis] |
| 18 |
Hb_000031_040 |
0.3571600103 |
- |
- |
centromere protein O [Medicago truncatula] |
| 19 |
Hb_000069_680 |
0.3601258016 |
- |
- |
- |
| 20 |
Hb_019093_010 |
0.3610113643 |
- |
- |
PREDICTED: uncharacterized protein LOC105766879 [Gossypium raimondii] |