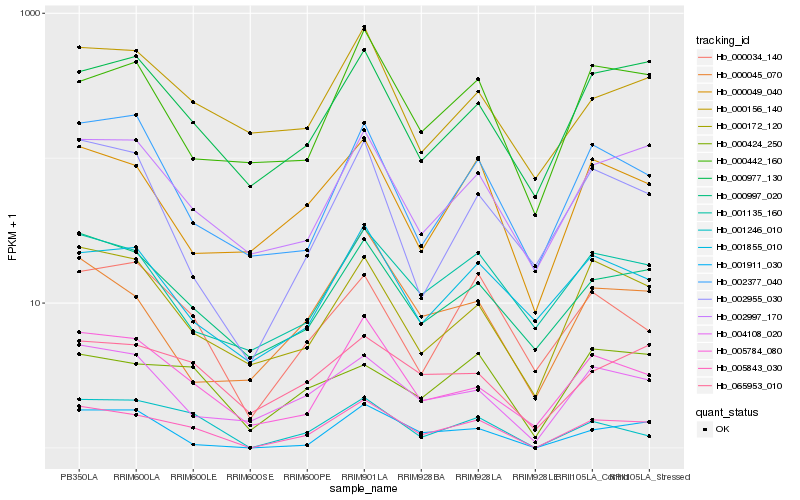
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005843_030 |
0.0 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 2 |
Hb_001246_010 |
0.1459179869 |
- |
- |
- |
| 3 |
Hb_005784_080 |
0.1586632668 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_000997_020 |
0.1675433061 |
- |
- |
PREDICTED: uncharacterized protein LOC105629694 [Jatropha curcas] |
| 5 |
Hb_002997_170 |
0.1740319593 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 6 |
Hb_004108_020 |
0.1752696841 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of NFKB 1 [Populus euphratica] |
| 7 |
Hb_000172_120 |
0.1753943456 |
- |
- |
PREDICTED: uncharacterized protein LOC105650770 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000977_130 |
0.1809630415 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm6 [Populus euphratica] |
| 9 |
Hb_065953_010 |
0.1814261752 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 10 |
Hb_000049_040 |
0.1826445789 |
- |
- |
PREDICTED: uncharacterized protein LOC105644475 [Jatropha curcas] |
| 11 |
Hb_000034_140 |
0.1834277036 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_001911_030 |
0.1839213879 |
- |
- |
PREDICTED: disease resistance RPP8-like protein 3 [Jatropha curcas] |
| 13 |
Hb_002377_040 |
0.1898017722 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000424_250 |
0.1898078999 |
- |
- |
PREDICTED: DNA cross-link repair protein SNM1 [Jatropha curcas] |
| 15 |
Hb_002955_030 |
0.1900625726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001135_160 |
0.1903566937 |
- |
- |
PREDICTED: elongator complex protein 6 [Jatropha curcas] |
| 17 |
Hb_000156_140 |
0.1913088212 |
- |
- |
Ribosomal protein L18ae/LX family protein [Theobroma cacao] |
| 18 |
Hb_000045_070 |
0.1918789711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001855_010 |
0.1919645094 |
- |
- |
PREDICTED: uncharacterized protein KIAA0930 homolog isoform X2 [Jatropha curcas] |
| 20 |
Hb_000442_160 |
0.1927586565 |
- |
- |
alpha-soluble nsf attachment protein, putative [Ricinus communis] |