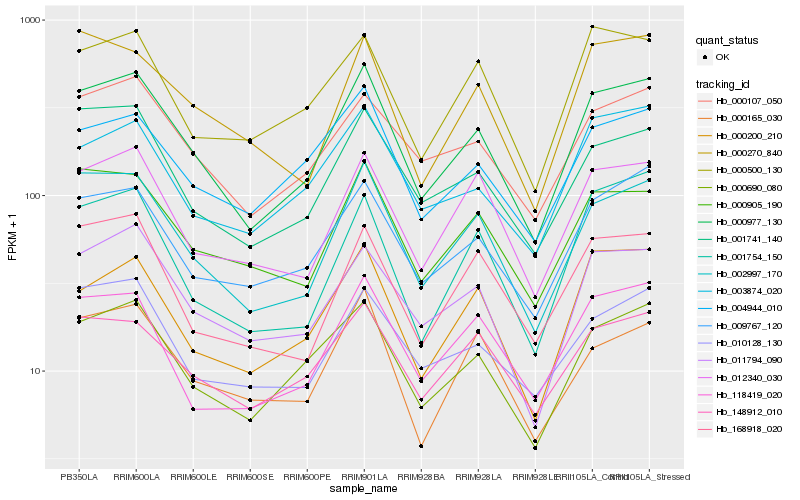
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_130 |
0.0 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm6 [Populus euphratica] |
| 2 |
Hb_002997_170 |
0.0553784881 |
- |
- |
PREDICTED: uncharacterized protein LOC105131887 [Populus euphratica] |
| 3 |
Hb_000690_080 |
0.0701323971 |
transcription factor |
TF Family: HB |
- |
| 4 |
Hb_001741_140 |
0.0808943293 |
- |
- |
PREDICTED: 60S ribosomal protein L7-2-like [Jatropha curcas] |
| 5 |
Hb_009767_120 |
0.0823340924 |
- |
- |
PREDICTED: probable prefoldin subunit 2 [Jatropha curcas] |
| 6 |
Hb_011794_090 |
0.0826969281 |
- |
- |
PREDICTED: uncharacterized protein LOC105642993 [Jatropha curcas] |
| 7 |
Hb_000107_050 |
0.0836178561 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 8 |
Hb_000200_210 |
0.0842895632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000905_190 |
0.085541529 |
- |
- |
PREDICTED: U6 snRNA-associated Sm-like protein LSm7 [Elaeis guineensis] |
| 10 |
Hb_012340_030 |
0.0885743504 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 6 [Jatropha curcas] |
| 11 |
Hb_004944_010 |
0.0896323781 |
- |
- |
small nuclear ribonucleoprotein E [Hevea brasiliensis] |
| 12 |
Hb_118419_020 |
0.0919616286 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Vitis vinifera] |
| 13 |
Hb_003874_020 |
0.0932052428 |
- |
- |
hypothetical protein POPTR_0012s02280g [Populus trichocarpa] |
| 14 |
Hb_010128_130 |
0.0947124902 |
- |
- |
PREDICTED: G patch domain-containing protein 8 isoform X1 [Jatropha curcas] |
| 15 |
Hb_148912_010 |
0.094724926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_168918_020 |
0.0948541011 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 17 |
Hb_000500_130 |
0.0963891592 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 18 |
Hb_001754_150 |
0.0972795428 |
- |
- |
PREDICTED: vesicle-associated membrane protein 724 [Jatropha curcas] |
| 19 |
Hb_000270_840 |
0.0974859672 |
- |
- |
PREDICTED: probable 26S proteasome complex subunit sem1-2 [Nicotiana tomentosiformis] |
| 20 |
Hb_000165_030 |
0.0975268158 |
- |
- |
PREDICTED: uncharacterized protein LOC105645673 isoform X1 [Jatropha curcas] |