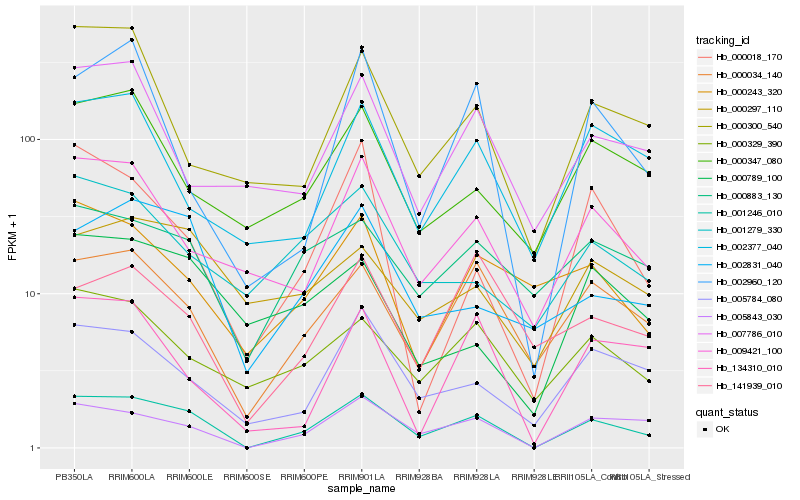
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001246_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_005843_030 |
0.1459179869 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 3 |
Hb_000034_140 |
0.1740642819 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic-like isoform X3 [Jatropha curcas] |
| 4 |
Hb_005784_080 |
0.1966685728 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_009421_100 |
0.1979393858 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0234100-like [Jatropha curcas] |
| 6 |
Hb_002831_040 |
0.2043644298 |
- |
- |
PREDICTED: fatty acid amide hydrolase-like [Jatropha curcas] |
| 7 |
Hb_000329_390 |
0.2043916465 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000883_130 |
0.2091309816 |
- |
- |
PREDICTED: novel plant SNARE 11 [Jatropha curcas] |
| 9 |
Hb_002960_120 |
0.2140251807 |
- |
- |
PREDICTED: uncharacterized protein LOC105635318 [Jatropha curcas] |
| 10 |
Hb_000018_170 |
0.2190675198 |
- |
- |
PREDICTED: bark storage protein A-like [Jatropha curcas] |
| 11 |
Hb_000347_080 |
0.2193019103 |
- |
- |
PREDICTED: 25.3 kDa vesicle transport protein [Jatropha curcas] |
| 12 |
Hb_134310_010 |
0.2195316851 |
- |
- |
PREDICTED: ATP-dependent DNA helicase MER3 homolog [Jatropha curcas] |
| 13 |
Hb_002377_040 |
0.2210370462 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 14 |
Hb_000300_540 |
0.2212094489 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 15 |
Hb_001279_330 |
0.2217797196 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 16 |
Hb_000297_110 |
0.2227437837 |
- |
- |
PREDICTED: uncharacterized protein LOC105634718 [Jatropha curcas] |
| 17 |
Hb_000243_320 |
0.2231607956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007786_010 |
0.2238755258 |
- |
- |
PREDICTED: uncharacterized protein LOC100801920 [Glycine max] |
| 19 |
Hb_000789_100 |
0.224295551 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 20 |
Hb_141939_010 |
0.2247640305 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 3, chloroplastic-like isoform X1 [Jatropha curcas] |