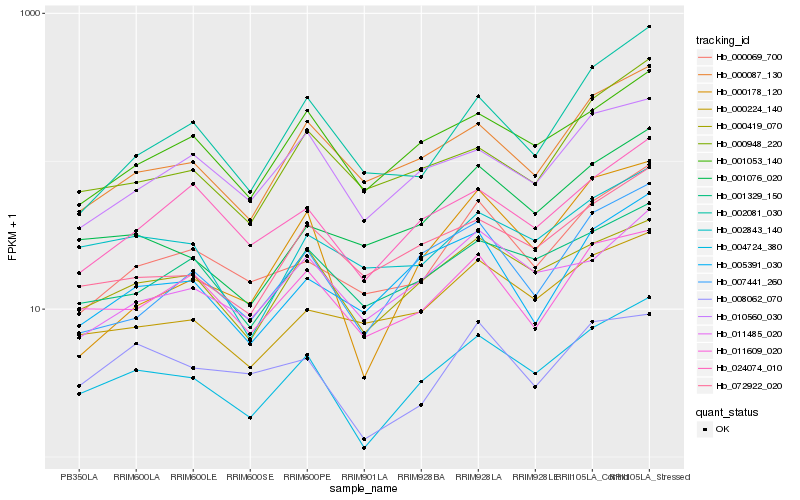
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004724_380 |
0.0 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein B-like [Jatropha curcas] |
| 2 |
Hb_000419_070 |
0.1331031911 |
- |
- |
V-type proton ATPase subunit G 1 [Jatropha curcas] |
| 3 |
Hb_007441_260 |
0.1365658982 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000087_130 |
0.1412198153 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001053_140 |
0.1437712376 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000069_700 |
0.1441800776 |
- |
- |
hypothetical protein B456_005G003200 [Gossypium raimondii] |
| 7 |
Hb_072922_020 |
0.1447894754 |
- |
- |
SNARE-like superfamily protein [Theobroma cacao] |
| 8 |
Hb_000178_120 |
0.1467003588 |
- |
- |
PREDICTED: histone H2B [Jatropha curcas] |
| 9 |
Hb_001329_150 |
0.1475644018 |
- |
- |
PREDICTED: autophagy-related protein 8i-like [Jatropha curcas] |
| 10 |
Hb_001076_020 |
0.1479275337 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 11 |
Hb_002843_140 |
0.1482867539 |
- |
- |
hypothetical protein JCGZ_20933 [Jatropha curcas] |
| 12 |
Hb_024074_010 |
0.1504469534 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 13 |
Hb_008062_070 |
0.151060283 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 14 |
Hb_011609_020 |
0.1521041586 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000224_140 |
0.1537943073 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 16 |
Hb_005391_030 |
0.1544174241 |
- |
- |
putative cytidine/deoxycytidylate deaminase family protein [Zea mays] |
| 17 |
Hb_011485_020 |
0.1557108279 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_010560_030 |
0.1557809065 |
- |
- |
PREDICTED: mitochondrial pyruvate carrier 4-like [Jatropha curcas] |
| 19 |
Hb_002081_030 |
0.1563242018 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000948_220 |
0.1564098899 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |