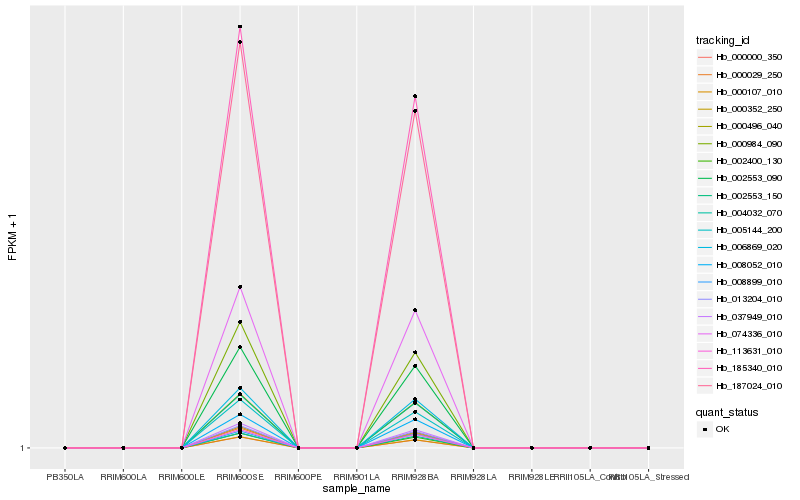
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 2 |
Hb_002553_090 |
0.001130007 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 3 |
Hb_006869_020 |
0.0031635851 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_008899_010 |
0.0087968174 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Nelumbo nucifera] |
| 5 |
Hb_185340_010 |
0.0100033871 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_074336_010 |
0.0102183655 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 7 |
Hb_187024_010 |
0.0104616996 |
- |
- |
- |
| 8 |
Hb_000496_040 |
0.0112758723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002553_150 |
0.0117365991 |
- |
- |
Gram-positive signal peptide protein, YSIRK family [Streptococcus pneumoniae TCH8431/19A] |
| 10 |
Hb_013204_010 |
0.0141029202 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 11 |
Hb_000352_250 |
0.0144372282 |
- |
- |
- |
| 12 |
Hb_008052_010 |
0.0147794191 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_000029_250 |
0.0148554867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_113631_010 |
0.0153706338 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_005144_200 |
0.0158470954 |
- |
- |
PREDICTED: uncharacterized protein LOC105642066 [Jatropha curcas] |
| 16 |
Hb_002400_130 |
0.0165082695 |
- |
- |
PREDICTED: uncharacterized protein LOC105637626 [Jatropha curcas] |
| 17 |
Hb_000984_090 |
0.0170360657 |
- |
- |
- |
| 18 |
Hb_037949_010 |
0.0172044154 |
- |
- |
hypothetical protein JCGZ_11175 [Jatropha curcas] |
| 19 |
Hb_000107_010 |
0.0176361301 |
- |
- |
- |
| 20 |
Hb_000000_350 |
0.0176867639 |
- |
- |
- |