| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
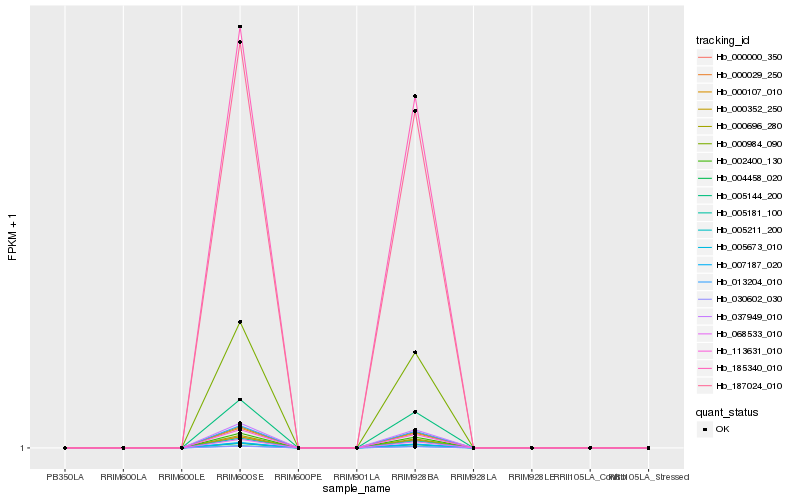
Hb_000352_250 |
0.0 |
- |
- |
- |
| 2 |
Hb_013204_010 |
0.0003343325 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 3 |
Hb_000029_250 |
0.0004182909 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_113631_010 |
0.0009334803 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_005144_200 |
0.0014099836 |
- |
- |
PREDICTED: uncharacterized protein LOC105642066 [Jatropha curcas] |
| 6 |
Hb_002400_130 |
0.0020712195 |
- |
- |
PREDICTED: uncharacterized protein LOC105637626 [Jatropha curcas] |
| 7 |
Hb_000984_090 |
0.0025990683 |
- |
- |
- |
| 8 |
Hb_037949_010 |
0.0027674353 |
- |
- |
hypothetical protein JCGZ_11175 [Jatropha curcas] |
| 9 |
Hb_000107_010 |
0.003199196 |
- |
- |
- |
| 10 |
Hb_000000_350 |
0.0032498353 |
- |
- |
- |
| 11 |
Hb_005181_100 |
0.0036563318 |
- |
- |
hypothetical protein POPTR_0010s15550g [Populus trichocarpa] |
| 12 |
Hb_187024_010 |
0.0039757447 |
- |
- |
- |
| 13 |
Hb_185340_010 |
0.0044340716 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_000696_280 |
0.0047124612 |
- |
- |
protein phosphatase-2c, putative [Ricinus communis] |
| 15 |
Hb_007187_020 |
0.0049717258 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 16 |
Hb_005211_200 |
0.0051243008 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 17 |
Hb_004458_020 |
0.0052362433 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 18 |
Hb_068533_010 |
0.0053196436 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_005673_010 |
0.0059569266 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 20 |
Hb_030602_030 |
0.0060882531 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |