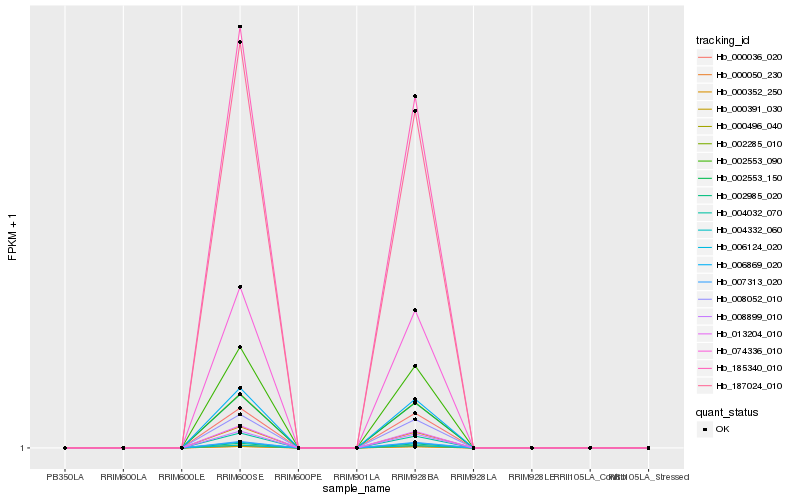
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006869_020 |
0.0 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_002553_090 |
0.0020335807 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 3 |
Hb_004032_070 |
0.0031635851 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 4 |
Hb_008899_010 |
0.0056332879 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Nelumbo nucifera] |
| 5 |
Hb_074336_010 |
0.0070548613 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 6 |
Hb_000496_040 |
0.0081123898 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002553_150 |
0.0085731269 |
- |
- |
Gram-positive signal peptide protein, YSIRK family [Streptococcus pneumoniae TCH8431/19A] |
| 8 |
Hb_008052_010 |
0.0116160264 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_185340_010 |
0.0131668233 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_187024_010 |
0.0136251235 |
- |
- |
- |
| 11 |
Hb_002285_010 |
0.0148982048 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_000050_230 |
0.0149083572 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_000391_030 |
0.0149416545 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 14 |
Hb_004332_060 |
0.0154652794 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 15 |
Hb_007313_020 |
0.0156994972 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000036_020 |
0.0163116231 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_002985_020 |
0.0163413247 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 18 |
Hb_006124_020 |
0.0168272387 |
- |
- |
hypothetical protein JCGZ_13738 [Jatropha curcas] |
| 19 |
Hb_013204_010 |
0.0172662296 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 20 |
Hb_000352_250 |
0.0176005256 |
- |
- |
- |