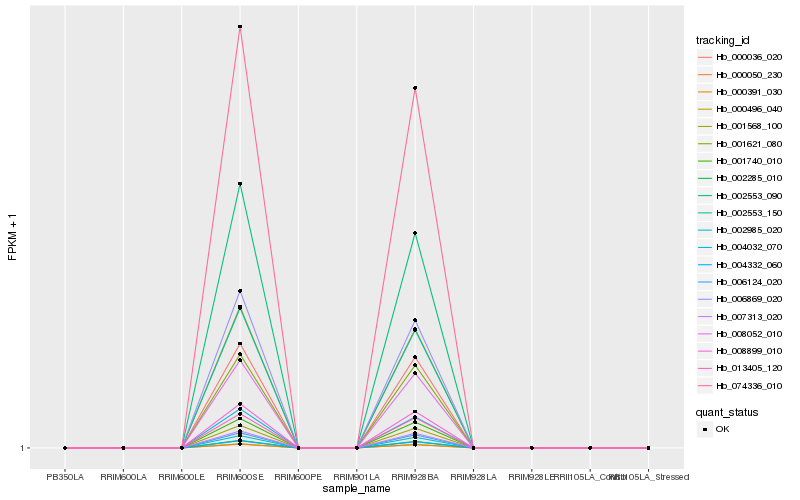
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008899_010 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 isoform X1 [Nelumbo nucifera] |
| 2 |
Hb_074336_010 |
0.0014215934 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 3 |
Hb_000496_040 |
0.0024791419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002553_150 |
0.0029398892 |
- |
- |
Gram-positive signal peptide protein, YSIRK family [Streptococcus pneumoniae TCH8431/19A] |
| 5 |
Hb_006869_020 |
0.0056332879 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_008052_010 |
0.0059828768 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_002553_090 |
0.0076668374 |
- |
- |
PREDICTED: uncharacterized protein LOC105628311 [Jatropha curcas] |
| 8 |
Hb_004032_070 |
0.0087968174 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_002285_010 |
0.0092651913 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_000050_230 |
0.0092753442 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_000391_030 |
0.009308643 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_004332_060 |
0.0098322937 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_007313_020 |
0.0100665233 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000036_020 |
0.0106786812 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002985_020 |
0.0107083844 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 16 |
Hb_006124_020 |
0.011194325 |
- |
- |
hypothetical protein JCGZ_13738 [Jatropha curcas] |
| 17 |
Hb_001568_100 |
0.0124782421 |
- |
- |
hypothetical protein Csa_2G404850 [Cucumis sativus] |
| 18 |
Hb_001740_010 |
0.0136683439 |
- |
- |
hypothetical protein JCGZ_18625 [Jatropha curcas] |
| 19 |
Hb_001621_080 |
0.0141719267 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 33 [Jatropha curcas] |
| 20 |
Hb_013405_120 |
0.0145253257 |
- |
- |
- |