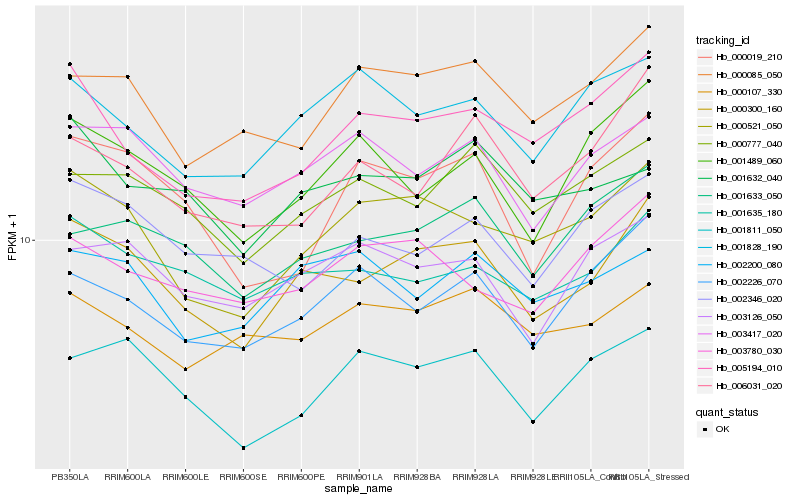
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_160 |
0.0 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: cyclic dof factor 3-like [Jatropha curcas] |
| 2 |
Hb_000521_050 |
0.0897619443 |
- |
- |
PREDICTED: major facilitator superfamily domain-containing protein 12-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_001635_180 |
0.0949481121 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At5g53840-like [Jatropha curcas] |
| 4 |
Hb_003417_020 |
0.1045060692 |
- |
- |
PREDICTED: 50S ribosomal protein L13, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001633_050 |
0.1050278804 |
- |
- |
PREDICTED: histone H2A.Z-specific chaperone CHZ1 [Jatropha curcas] |
| 6 |
Hb_003126_050 |
0.1069140881 |
- |
- |
PREDICTED: cell cycle checkpoint protein RAD17 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000777_040 |
0.1074361448 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 8 |
Hb_005194_010 |
0.1079479918 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 9 |
Hb_002226_070 |
0.1117989289 |
- |
- |
PREDICTED: uncharacterized CRM domain-containing protein At3g25440, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000085_050 |
0.1123897646 |
- |
- |
PREDICTED: grpE protein homolog, mitochondrial isoform X1 [Jatropha curcas] |
| 11 |
Hb_001489_060 |
0.1140478957 |
- |
- |
PREDICTED: uncharacterized protein LOC105649778 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000019_210 |
0.1157278121 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 13 |
Hb_003780_030 |
0.1158590067 |
- |
- |
PREDICTED: protein CMSS1 [Jatropha curcas] |
| 14 |
Hb_002200_080 |
0.1176655199 |
- |
- |
PREDICTED: uncharacterized protein C57A10.07 [Jatropha curcas] |
| 15 |
Hb_002346_020 |
0.1187217014 |
- |
- |
- |
| 16 |
Hb_001828_190 |
0.1189110372 |
- |
- |
PREDICTED: phosducin-like protein 3 [Prunus mume] |
| 17 |
Hb_001811_050 |
0.1189166248 |
- |
- |
PREDICTED: protein EXECUTER 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_006031_020 |
0.1190017653 |
- |
- |
PREDICTED: protein FLX-like 1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000107_330 |
0.1192150744 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38150 [Jatropha curcas] |
| 20 |
Hb_001632_040 |
0.1192491917 |
- |
- |
PREDICTED: transmembrane protein adipocyte-associated 1 homolog isoform X1 [Jatropha curcas] |