| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
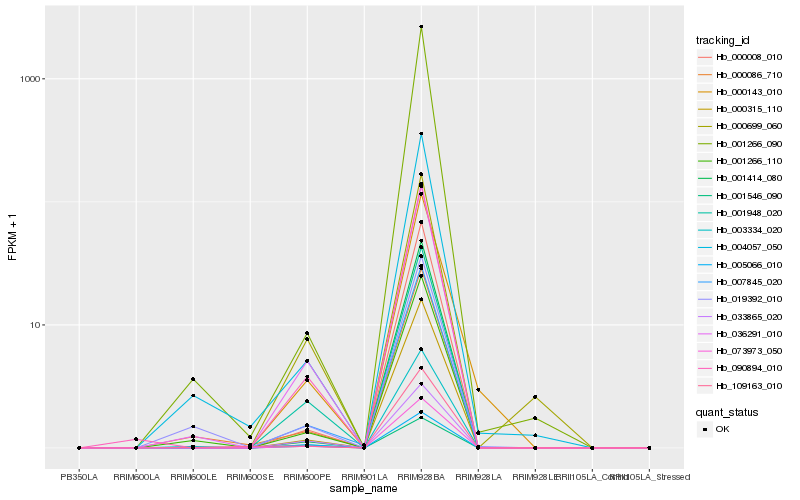
Hb_000143_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 2 |
Hb_073973_050 |
0.0914277345 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 2 protein [Jatropha curcas] |
| 3 |
Hb_090894_010 |
0.0946188632 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 4 |
Hb_001948_020 |
0.0953976578 |
- |
- |
- |
| 5 |
Hb_007845_020 |
0.0965621014 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 6 |
Hb_036291_010 |
0.0983477051 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 7 |
Hb_000315_110 |
0.0988033675 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 1-like [Jatropha curcas] |
| 8 |
Hb_001546_090 |
0.1025774871 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB114-like [Jatropha curcas] |
| 9 |
Hb_004057_050 |
0.1034141608 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 10 |
Hb_000008_010 |
0.1084097556 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 11 |
Hb_001266_110 |
0.1093533726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003334_020 |
0.1104668847 |
- |
- |
hypothetical protein CISIN_1g041546mg [Citrus sinensis] |
| 13 |
Hb_019392_010 |
0.117747693 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 14 |
Hb_000086_710 |
0.1192784881 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 15 |
Hb_033865_020 |
0.1197094369 |
- |
- |
- |
| 16 |
Hb_001266_090 |
0.120065851 |
- |
- |
- |
| 17 |
Hb_001414_080 |
0.1203687901 |
- |
- |
PREDICTED: probable S-adenosylmethionine-dependent methyltransferase At5g38100 [Jatropha curcas] |
| 18 |
Hb_000699_060 |
0.1207758902 |
- |
- |
hypothetical protein POPTR_0019s03180g [Populus trichocarpa] |
| 19 |
Hb_005066_010 |
0.1221817159 |
- |
- |
FAD-binding domain-containing family protein [Populus trichocarpa] |
| 20 |
Hb_109163_010 |
0.123943051 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |