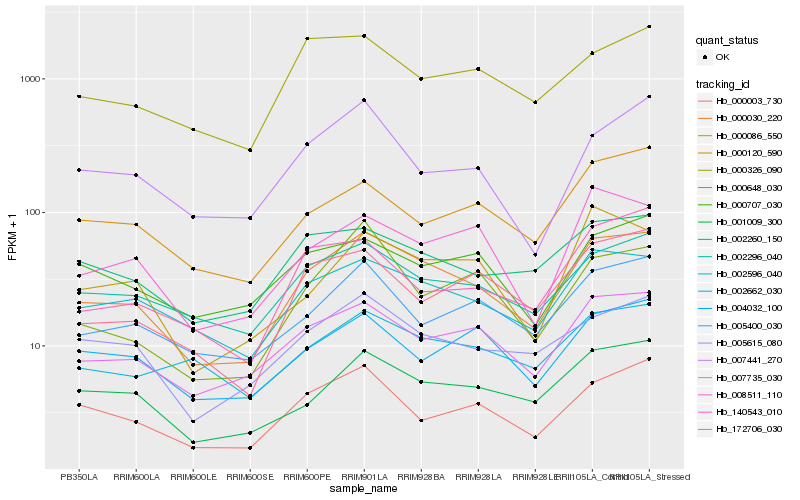
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_220 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650405 [Jatropha curcas] |
| 2 |
Hb_007735_030 |
0.0943728353 |
- |
- |
Calcium-binding EF-hand family protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_002596_040 |
0.0972615071 |
- |
- |
PREDICTED: uncharacterized protein LOC105645264 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001009_300 |
0.1046022027 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3-like [Gossypium raimondii] |
| 5 |
Hb_002296_040 |
0.1056480575 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 4 [Jatropha curcas] |
| 6 |
Hb_005615_080 |
0.108855533 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |
| 7 |
Hb_000003_730 |
0.1141427844 |
- |
- |
DNA repair/transcription protein met18/mms19, putative [Ricinus communis] |
| 8 |
Hb_002260_150 |
0.1168439884 |
- |
- |
PREDICTED: uncharacterized protein LOC105647187 isoform X2 [Jatropha curcas] |
| 9 |
Hb_005400_030 |
0.1191122875 |
- |
- |
PREDICTED: LYR motif-containing protein At3g19508 [Jatropha curcas] |
| 10 |
Hb_008511_110 |
0.1227532779 |
- |
- |
PREDICTED: uncharacterized protein LOC105134715 [Populus euphratica] |
| 11 |
Hb_172706_030 |
0.1236078155 |
- |
- |
PREDICTED: uncharacterized protein LOC105640907 [Jatropha curcas] |
| 12 |
Hb_000326_090 |
0.1262944508 |
- |
- |
PREDICTED: glycine-rich RNA-binding protein GRP1A-like [Nelumbo nucifera] |
| 13 |
Hb_002662_030 |
0.1266728379 |
- |
- |
PREDICTED: mannose-P-dolichol utilization defect 1 protein homolog 2 [Jatropha curcas] |
| 14 |
Hb_000648_030 |
0.1267870592 |
- |
- |
hypothetical protein PHAVU_002G055100g [Phaseolus vulgaris] |
| 15 |
Hb_000120_590 |
0.1269593011 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 16 |
Hb_000086_550 |
0.1269903225 |
- |
- |
Os02g0814700 [Oryza sativa Japonica Group] |
| 17 |
Hb_004032_100 |
0.1281171471 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF3-like [Jatropha curcas] |
| 18 |
Hb_007441_270 |
0.1286394209 |
- |
- |
PREDICTED: 40S ribosomal protein S11 [Jatropha curcas] |
| 19 |
Hb_000707_030 |
0.1293664699 |
- |
- |
PREDICTED: probable proteasome inhibitor [Vitis vinifera] |
| 20 |
Hb_140543_010 |
0.1307252931 |
- |
- |
- |