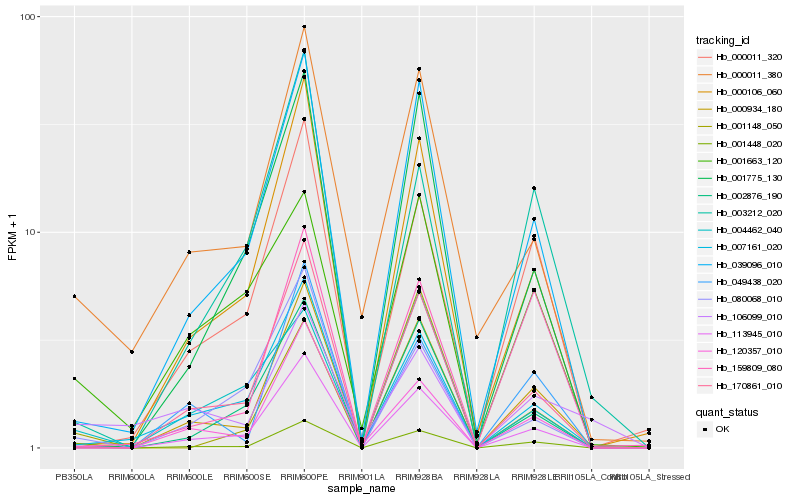
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001448_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_17204 [Jatropha curcas] |
| 2 |
Hb_113945_010 |
0.1768140002 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000934_180 |
0.1928027189 |
- |
- |
hypothetical protein JCGZ_25237 [Jatropha curcas] |
| 4 |
Hb_039096_010 |
0.1989246897 |
- |
- |
unknown [Populus trichocarpa] |
| 5 |
Hb_000011_380 |
0.2051372155 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 6 |
Hb_000106_060 |
0.2125646115 |
- |
- |
PREDICTED: uncharacterized protein LOC105636580 [Jatropha curcas] |
| 7 |
Hb_170861_010 |
0.2157595003 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 8 |
Hb_002876_190 |
0.2218417747 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 9 |
Hb_007161_020 |
0.22902029 |
- |
- |
PREDICTED: serine/threonine-protein kinase BRI1-like 2 [Jatropha curcas] |
| 10 |
Hb_001663_120 |
0.2296013504 |
- |
- |
PREDICTED: formin-like protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_120357_010 |
0.2308093001 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000011_320 |
0.233363447 |
- |
- |
PREDICTED: uncharacterized protein LOC105631328 [Jatropha curcas] |
| 13 |
Hb_001775_130 |
0.234330776 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 14 |
Hb_080068_010 |
0.2345106234 |
- |
- |
PREDICTED: basic 7S globulin-like [Jatropha curcas] |
| 15 |
Hb_049438_020 |
0.2379003356 |
- |
- |
PREDICTED: peroxidase 20 [Jatropha curcas] |
| 16 |
Hb_159809_080 |
0.2402968297 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_004462_040 |
0.2412771917 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001148_050 |
0.2440854245 |
- |
- |
- |
| 19 |
Hb_106099_010 |
0.2455494789 |
- |
- |
- |
| 20 |
Hb_003212_020 |
0.2458651199 |
- |
- |
hypothetical protein PHAVU_009G114800g [Phaseolus vulgaris] |