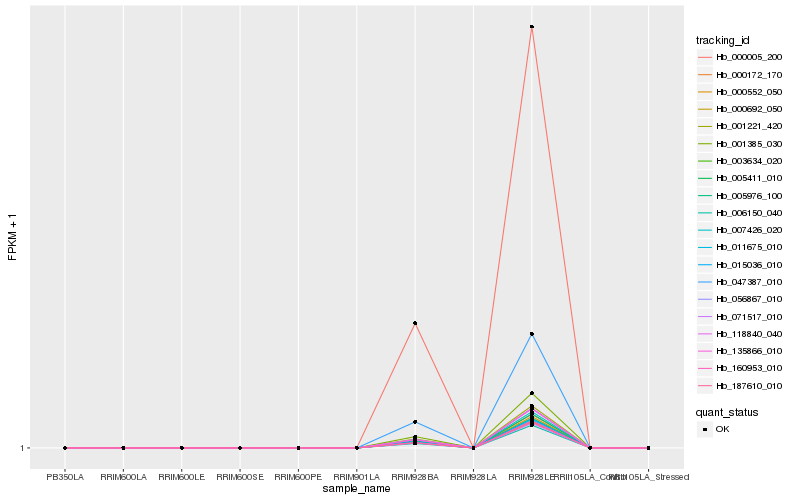
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000692_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_005976_100 |
0.0001124593 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein RCOM_1408460 [Ricinus communis] |
| 3 |
Hb_000552_050 |
0.0010353532 |
- |
- |
- |
| 4 |
Hb_007426_020 |
0.001110412 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 5 |
Hb_005411_010 |
0.0017557739 |
- |
- |
- |
| 6 |
Hb_011675_010 |
0.0019917544 |
- |
- |
- |
| 7 |
Hb_001385_030 |
0.0023478707 |
- |
- |
PREDICTED: uncharacterized protein LOC100852852 [Vitis vinifera] |
| 8 |
Hb_015036_010 |
0.0023479875 |
- |
- |
- |
| 9 |
Hb_135866_010 |
0.0024713841 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 10 |
Hb_118840_040 |
0.002630133 |
- |
- |
- |
| 11 |
Hb_056867_010 |
0.0027105185 |
- |
- |
PREDICTED: uncharacterized protein LOC105786773 [Gossypium raimondii] |
| 12 |
Hb_160953_010 |
0.0029295558 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 13 |
Hb_003634_020 |
0.0030317702 |
- |
- |
PREDICTED: uncharacterized protein LOC105775392, partial [Gossypium raimondii] |
| 14 |
Hb_187610_010 |
0.0033594309 |
- |
- |
PREDICTED: uncharacterized protein LOC105785094 [Gossypium raimondii] |
| 15 |
Hb_000172_170 |
0.0035148417 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_000005_200 |
0.0037117378 |
- |
- |
- |
| 17 |
Hb_047387_010 |
0.0037380399 |
- |
- |
- |
| 18 |
Hb_001221_420 |
0.003829895 |
- |
- |
Uncharacterized protein TCM_032186 [Theobroma cacao] |
| 19 |
Hb_071517_010 |
0.0039571727 |
- |
- |
hypothetical protein CICLE_v10010288mg, partial [Citrus clementina] |
| 20 |
Hb_006150_040 |
0.0047323353 |
- |
- |
PREDICTED: uncharacterized protein LOC105772114 [Gossypium raimondii] |