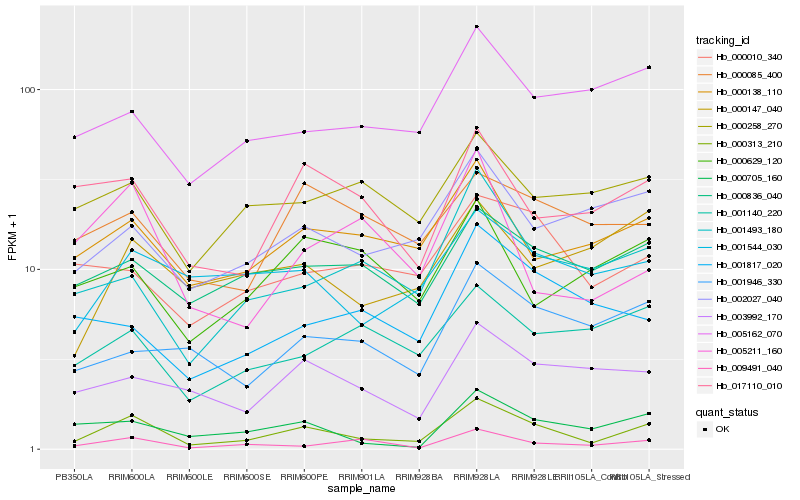
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000313_210 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_009491_040 |
0.2003843984 |
- |
- |
PREDICTED: uncharacterized protein LOC105636967 [Jatropha curcas] |
| 3 |
Hb_000705_160 |
0.2029631847 |
- |
- |
hypothetical protein CISIN_1g042743mg [Citrus sinensis] |
| 4 |
Hb_001493_180 |
0.2038347181 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 5 |
Hb_000138_110 |
0.2065154547 |
- |
- |
PREDICTED: probable protein phosphatase 2C 68 [Jatropha curcas] |
| 6 |
Hb_017110_010 |
0.2098468366 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g55760 [Jatropha curcas] |
| 7 |
Hb_002027_040 |
0.2126248719 |
- |
- |
synapse-associated protein, putative [Ricinus communis] |
| 8 |
Hb_003992_170 |
0.2168608266 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |
| 9 |
Hb_000836_040 |
0.2169501675 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001544_030 |
0.2192691589 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 11 |
Hb_005162_070 |
0.219603667 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000010_340 |
0.2222789481 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005211_160 |
0.2249103822 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 14 |
Hb_000147_040 |
0.2262687418 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 15 |
Hb_001946_330 |
0.2281275622 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 16 |
Hb_001140_220 |
0.2289841214 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000258_270 |
0.2298656148 |
- |
- |
PREDICTED: cullin-3A [Jatropha curcas] |
| 18 |
Hb_000085_400 |
0.2314573166 |
- |
- |
Heterogeneous nuclear ribonucleoprotein 27C, putative [Ricinus communis] |
| 19 |
Hb_001817_020 |
0.2325984572 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 20 |
Hb_000629_120 |
0.2337242944 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein [Theobroma cacao] |