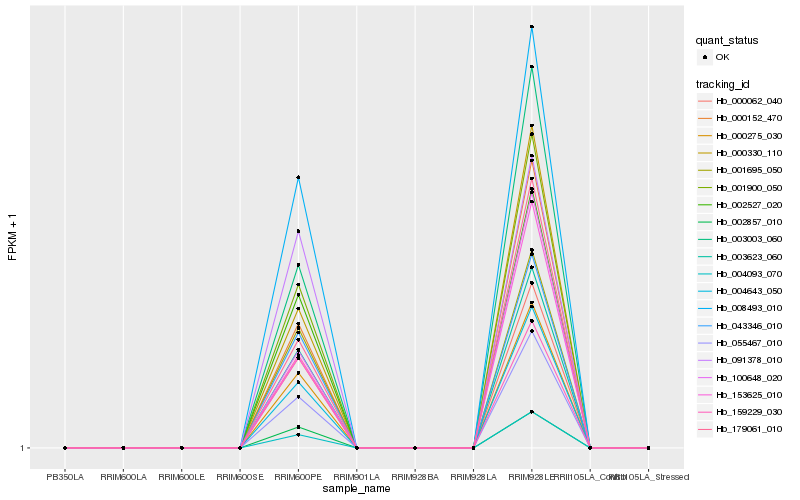
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000275_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001900_050 |
0.0036077681 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 3 |
Hb_003623_060 |
0.0100614349 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000062_040 |
0.0109743826 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 5 |
Hb_004643_050 |
0.0207381205 |
- |
- |
hypothetical protein B456_001G165500 [Gossypium raimondii] |
| 6 |
Hb_003003_060 |
0.0231274367 |
- |
- |
- |
| 7 |
Hb_002527_020 |
0.0249740937 |
- |
- |
PREDICTED: F-box protein At1g30790-like [Jatropha curcas] |
| 8 |
Hb_002857_010 |
0.0264926727 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 9 |
Hb_043346_010 |
0.0287046148 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |
| 10 |
Hb_001695_050 |
0.031628265 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_055467_010 |
0.0318920711 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 12 |
Hb_008493_010 |
0.0387015638 |
- |
- |
hypothetical protein JCGZ_10529 [Jatropha curcas] |
| 13 |
Hb_000152_470 |
0.0409274737 |
- |
- |
- |
| 14 |
Hb_000330_110 |
0.0424362229 |
- |
- |
- |
| 15 |
Hb_179061_010 |
0.0547963174 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 16 |
Hb_004093_070 |
0.0629762246 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 17 |
Hb_100648_020 |
0.0650357108 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 18 |
Hb_153625_010 |
0.0714560387 |
- |
- |
PREDICTED: 21 kDa protein [Populus euphratica] |
| 19 |
Hb_159229_030 |
0.0734062125 |
- |
- |
hypothetical protein CISIN_1g0034891mg, partial [Citrus sinensis] |
| 20 |
Hb_091378_010 |
0.0740628586 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |