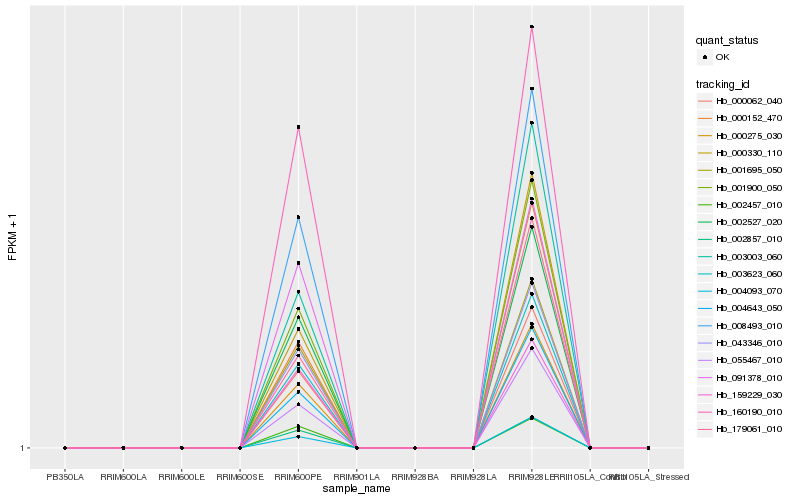
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 2 |
Hb_003623_060 |
0.0009129903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000275_030 |
0.0109743826 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002527_020 |
0.0140013105 |
- |
- |
PREDICTED: F-box protein At1g30790-like [Jatropha curcas] |
| 5 |
Hb_001900_050 |
0.0145819035 |
- |
- |
PREDICTED: uncharacterized protein LOC102609547 [Citrus sinensis] |
| 6 |
Hb_002857_010 |
0.0155201683 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 7 |
Hb_043346_010 |
0.0177325526 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |
| 8 |
Hb_001695_050 |
0.0206568529 |
- |
- |
PREDICTED: uncharacterized protein LOC104884077 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_008493_010 |
0.0277320302 |
- |
- |
hypothetical protein JCGZ_10529 [Jatropha curcas] |
| 10 |
Hb_004643_050 |
0.0317093598 |
- |
- |
hypothetical protein B456_001G165500 [Gossypium raimondii] |
| 11 |
Hb_003003_060 |
0.0340980407 |
- |
- |
- |
| 12 |
Hb_055467_010 |
0.0428598452 |
- |
- |
hypothetical protein POPTR_0009s13360g [Populus trichocarpa] |
| 13 |
Hb_000152_470 |
0.0518914915 |
- |
- |
- |
| 14 |
Hb_000330_110 |
0.0533995288 |
- |
- |
- |
| 15 |
Hb_159229_030 |
0.0624519401 |
- |
- |
hypothetical protein CISIN_1g0034891mg, partial [Citrus sinensis] |
| 16 |
Hb_091378_010 |
0.0631089686 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 17 |
Hb_002457_010 |
0.0643824188 |
- |
- |
polyprotein [Oryza australiensis] |
| 18 |
Hb_160190_010 |
0.0655138625 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 19 |
Hb_179061_010 |
0.065752855 |
- |
- |
hypothetical protein JCGZ_01311 [Jatropha curcas] |
| 20 |
Hb_004093_070 |
0.0739273433 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |