| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
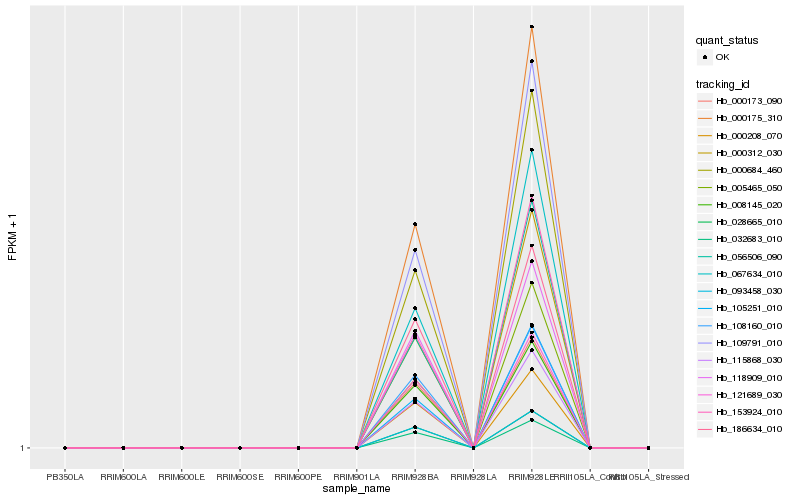
Hb_000175_310 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_109791_010 |
0.0066423016 |
- |
- |
- |
| 3 |
Hb_000684_460 |
0.012006513 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 4 |
Hb_115868_030 |
0.012895678 |
- |
- |
Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial -like protein [Gossypium arboreum] |
| 5 |
Hb_032683_010 |
0.0211130675 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 6 |
Hb_067634_010 |
0.0221411883 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 7 |
Hb_028665_010 |
0.0222263182 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 8 |
Hb_093458_030 |
0.0222347709 |
- |
- |
Cellulose synthase A catalytic subunit 1 [UDP-forming], putative [Ricinus communis] |
| 9 |
Hb_000312_030 |
0.022416226 |
- |
- |
PREDICTED: uncharacterized protein LOC104888072 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000208_070 |
0.0273538995 |
- |
- |
- |
| 11 |
Hb_153924_010 |
0.029359399 |
- |
- |
hypothetical protein JCGZ_22941 [Jatropha curcas] |
| 12 |
Hb_056506_090 |
0.0301106999 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 13 |
Hb_008145_020 |
0.0310021672 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 14 |
Hb_000173_090 |
0.0315109946 |
- |
- |
- |
| 15 |
Hb_121689_030 |
0.0321699924 |
- |
- |
hypothetical protein, partial [Staphylococcus aureus] |
| 16 |
Hb_108160_010 |
0.0330710408 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 17 |
Hb_005465_050 |
0.0417937267 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 18 |
Hb_118909_010 |
0.0422048567 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 19 |
Hb_186634_010 |
0.0445398842 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 20 |
Hb_105251_010 |
0.0472451919 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |