| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
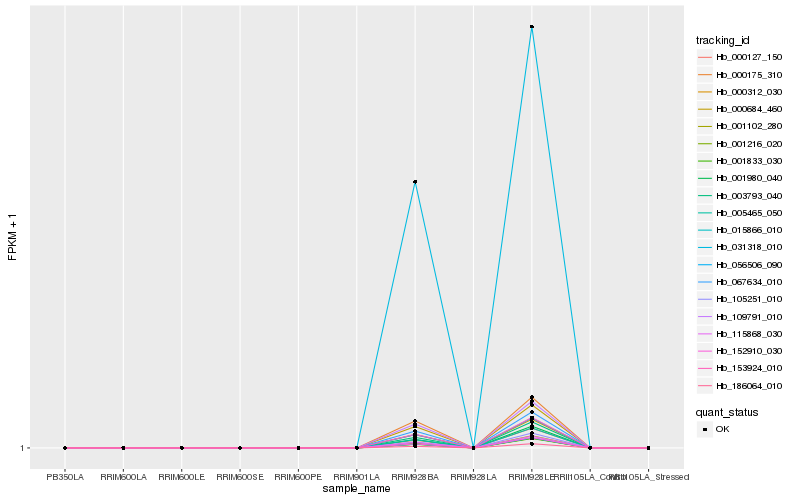
Hb_000312_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104888072 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_067634_010 |
0.0002750998 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 3 |
Hb_153924_010 |
0.0069452669 |
- |
- |
hypothetical protein JCGZ_22941 [Jatropha curcas] |
| 4 |
Hb_056506_090 |
0.0076968558 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 5 |
Hb_115868_030 |
0.0095217866 |
- |
- |
Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial -like protein [Gossypium arboreum] |
| 6 |
Hb_000684_460 |
0.0104109726 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 7 |
Hb_109791_010 |
0.0157749743 |
- |
- |
- |
| 8 |
Hb_005465_050 |
0.0193859373 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 9 |
Hb_000175_310 |
0.022416226 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 10 |
Hb_105251_010 |
0.0248412626 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003793_040 |
0.0262958983 |
- |
- |
hypothetical protein CARUB_v10021783mg [Capsella rubella] |
| 12 |
Hb_001102_280 |
0.0276146368 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 13 |
Hb_031318_010 |
0.029437791 |
- |
- |
- |
| 14 |
Hb_152910_030 |
0.0294403146 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 15 |
Hb_000127_150 |
0.0296908828 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Jatropha curcas] |
| 16 |
Hb_015866_010 |
0.0297989969 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 17 |
Hb_001833_030 |
0.0299475928 |
- |
- |
polyprotein [Oryza australiensis] |
| 18 |
Hb_001216_020 |
0.0303668366 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 19 |
Hb_001980_040 |
0.0321988779 |
- |
- |
- |
| 20 |
Hb_186064_010 |
0.0352885187 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |