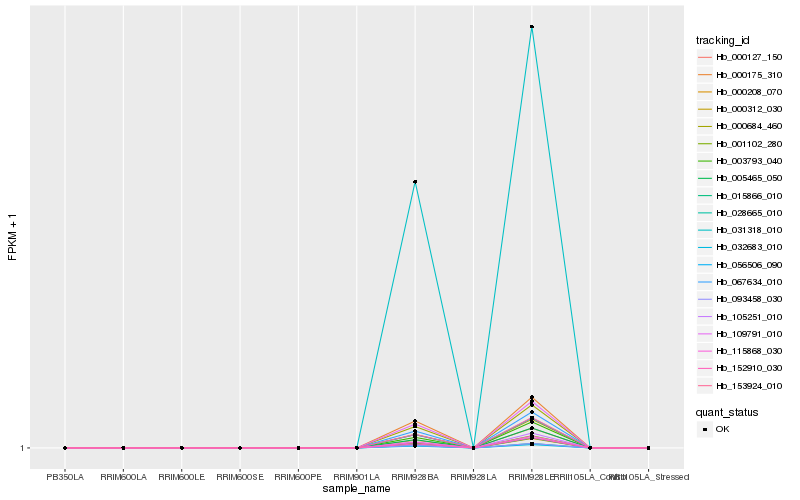
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_460 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 2 |
Hb_115868_030 |
0.0008892262 |
- |
- |
Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial -like protein [Gossypium arboreum] |
| 3 |
Hb_109791_010 |
0.0053644005 |
- |
- |
- |
| 4 |
Hb_067634_010 |
0.0101358862 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |
| 5 |
Hb_000312_030 |
0.0104109726 |
- |
- |
PREDICTED: uncharacterized protein LOC104888072 [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_000175_310 |
0.012006513 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_153924_010 |
0.0173556569 |
- |
- |
hypothetical protein JCGZ_22941 [Jatropha curcas] |
| 8 |
Hb_056506_090 |
0.0181071542 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 9 |
Hb_005465_050 |
0.029794078 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 10 |
Hb_032683_010 |
0.0331159719 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 11 |
Hb_028665_010 |
0.0342289089 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 12 |
Hb_093458_030 |
0.0342373591 |
- |
- |
Cellulose synthase A catalytic subunit 1 [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_105251_010 |
0.0352479149 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_003793_040 |
0.0367021009 |
- |
- |
hypothetical protein CARUB_v10021783mg [Capsella rubella] |
| 15 |
Hb_001102_280 |
0.0380204123 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_000208_070 |
0.0393548901 |
- |
- |
- |
| 17 |
Hb_031318_010 |
0.0398429454 |
- |
- |
- |
| 18 |
Hb_152910_030 |
0.0398454681 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 19 |
Hb_000127_150 |
0.0400959481 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Jatropha curcas] |
| 20 |
Hb_015866_010 |
0.040204024 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |