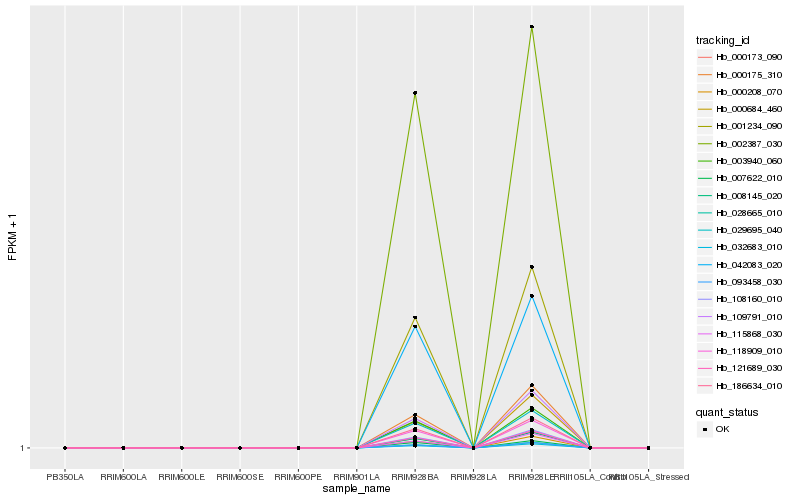
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_121689_030 |
0.0 |
- |
- |
hypothetical protein, partial [Staphylococcus aureus] |
| 2 |
Hb_000173_090 |
0.0006592709 |
- |
- |
- |
| 3 |
Hb_108160_010 |
0.0009014398 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 4 |
Hb_008145_020 |
0.0011683016 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000208_070 |
0.0048178318 |
- |
- |
- |
| 6 |
Hb_093458_030 |
0.0099381505 |
- |
- |
Cellulose synthase A catalytic subunit 1 [UDP-forming], putative [Ricinus communis] |
| 7 |
Hb_028665_010 |
0.0099466045 |
- |
- |
PREDICTED: uncharacterized protein LOC105803109 [Gossypium raimondii] |
| 8 |
Hb_118909_010 |
0.0100403847 |
- |
- |
PREDICTED: uncharacterized protein LOC102588546 [Solanum tuberosum] |
| 9 |
Hb_032683_010 |
0.0110600231 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_186634_010 |
0.0123770593 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 11 |
Hb_001234_090 |
0.0192740043 |
- |
- |
hypothetical protein CISIN_1g013527mg [Citrus sinensis] |
| 12 |
Hb_029695_040 |
0.0201969011 |
- |
- |
Pectate lyase precursor, putative [Ricinus communis] |
| 13 |
Hb_003940_060 |
0.0231613693 |
- |
- |
PREDICTED: uncharacterized protein LOC105784680 [Gossypium raimondii] |
| 14 |
Hb_000175_310 |
0.0321699924 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_109791_010 |
0.0388087852 |
- |
- |
- |
| 16 |
Hb_002387_030 |
0.0402317713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000684_460 |
0.04416925 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 18 |
Hb_115868_030 |
0.0450577141 |
- |
- |
Hexaprenyldihydroxybenzoate methyltransferase, mitochondrial -like protein [Gossypium arboreum] |
| 19 |
Hb_042083_020 |
0.0502675639 |
- |
- |
- |
| 20 |
Hb_007622_010 |
0.0511400977 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |