| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
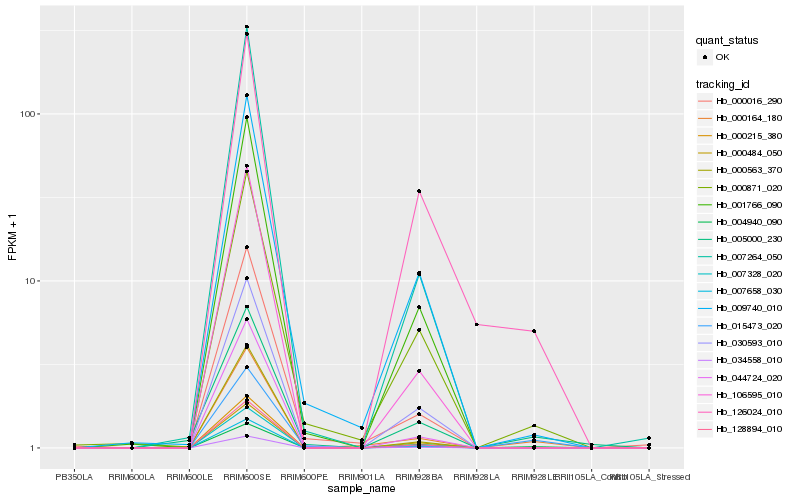
Hb_000164_180 |
0.0 |
- |
- |
(+)-delta-cadinene synthase isozyme A, putative [Ricinus communis] |
| 2 |
Hb_000871_020 |
0.129115626 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_015473_020 |
0.1334789848 |
- |
- |
hypothetical protein B456_001G090400 [Gossypium raimondii] |
| 4 |
Hb_009740_010 |
0.137562216 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_128894_010 |
0.1392076017 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_106595_010 |
0.1399497942 |
- |
- |
hypothetical protein B456_003G065300 [Gossypium raimondii] |
| 7 |
Hb_034558_010 |
0.1401676467 |
- |
- |
- |
| 8 |
Hb_004940_090 |
0.140701872 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 9 |
Hb_044724_020 |
0.1416396046 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000215_380 |
0.1440707499 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 11 |
Hb_007328_020 |
0.144422288 |
- |
- |
- |
| 12 |
Hb_000563_370 |
0.1446331045 |
- |
- |
hypothetical protein JCGZ_18919 [Jatropha curcas] |
| 13 |
Hb_007264_050 |
0.1463236583 |
- |
- |
- |
| 14 |
Hb_001766_090 |
0.1467280756 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 15 |
Hb_005000_230 |
0.1481721729 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 16 |
Hb_126024_010 |
0.1491748524 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Populus euphratica] |
| 17 |
Hb_000016_290 |
0.1495233814 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 18 |
Hb_007658_030 |
0.1495565832 |
- |
- |
PREDICTED: lupeol synthase-like isoform X1 [Populus euphratica] |
| 19 |
Hb_000484_050 |
0.1523358607 |
- |
- |
hypothetical protein CICLE_v10028549mg [Citrus clementina] |
| 20 |
Hb_030593_010 |
0.1530094484 |
- |
- |
hypothetical protein JCGZ_06063 [Jatropha curcas] |