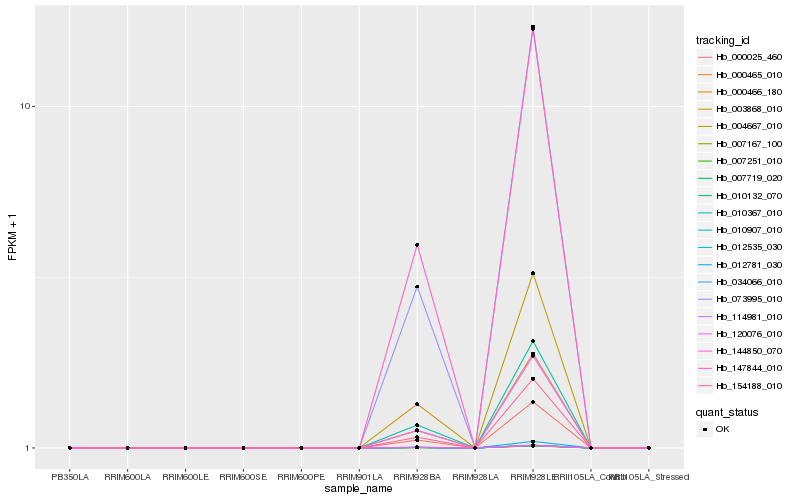
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_460 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104895905 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_120076_010 |
0.0017643964 |
- |
- |
PREDICTED: uncharacterized protein LOC104783951 [Camelina sativa] |
| 3 |
Hb_003868_010 |
0.004292792 |
- |
- |
- |
| 4 |
Hb_010367_010 |
0.008578561 |
- |
- |
- |
| 5 |
Hb_000465_010 |
0.010010262 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 1, mitochondrial-like [Citrus sinensis] |
| 6 |
Hb_154188_010 |
0.0237936627 |
- |
- |
hypothetical protein CICLE_v10003434mg, partial [Citrus clementina] |
| 7 |
Hb_012781_030 |
0.0264473278 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_073995_010 |
0.0273802245 |
- |
- |
receptor-like kinase [Medicago truncatula] |
| 9 |
Hb_000466_180 |
0.0313901047 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_007167_100 |
0.0315351558 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 11 |
Hb_007251_010 |
0.0317628285 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 12 |
Hb_010132_070 |
0.0318826548 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 13 |
Hb_034066_010 |
0.0319105936 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_144850_070 |
0.0319555415 |
- |
- |
OSJNBa0032N05.11 [Oryza sativa Japonica Group] |
| 15 |
Hb_012535_030 |
0.0319637294 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_114981_010 |
0.0320296641 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 17 |
Hb_004667_010 |
0.0320689473 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_007719_020 |
0.0320963174 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 19 |
Hb_010907_010 |
0.032171537 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 20 |
Hb_147844_010 |
0.0322658709 |
- |
- |
putative 60S ribosomal protein L37 [Aegilops tauschii] |