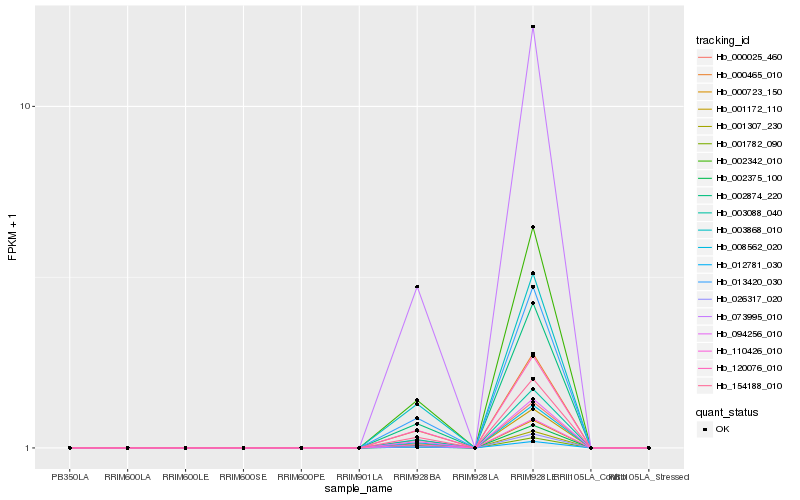
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073995_010 |
0.0 |
- |
- |
receptor-like kinase [Medicago truncatula] |
| 2 |
Hb_012781_030 |
0.0009338071 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_154188_010 |
0.0035896907 |
- |
- |
hypothetical protein CICLE_v10003434mg, partial [Citrus clementina] |
| 4 |
Hb_003088_040 |
0.0049867668 |
- |
- |
PREDICTED: uncharacterized protein LOC103417518 [Malus domestica] |
| 5 |
Hb_013420_030 |
0.0096770921 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_094256_010 |
0.012105348 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_002342_010 |
0.0130532377 |
- |
- |
RALF-LIKE 27 family protein [Populus trichocarpa] |
| 8 |
Hb_008562_020 |
0.0162380746 |
- |
- |
- |
| 9 |
Hb_002874_220 |
0.0168527513 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g31250 [Jatropha curcas] |
| 10 |
Hb_000465_010 |
0.0173761318 |
- |
- |
PREDICTED: 3-hydroxyisobutyryl-CoA hydrolase-like protein 1, mitochondrial-like [Citrus sinensis] |
| 11 |
Hb_110426_010 |
0.0240391476 |
- |
- |
- |
| 12 |
Hb_000723_150 |
0.0246851675 |
- |
- |
PREDICTED: high affinity nitrate transporter 2.4-like [Jatropha curcas] |
| 13 |
Hb_120076_010 |
0.0256174009 |
- |
- |
PREDICTED: uncharacterized protein LOC104783951 [Camelina sativa] |
| 14 |
Hb_002375_100 |
0.0271885642 |
- |
- |
PREDICTED: uncharacterized protein LOC104878944 [Vitis vinifera] |
| 15 |
Hb_000025_460 |
0.0273802245 |
- |
- |
PREDICTED: uncharacterized protein LOC104895905 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_001172_110 |
0.0280719985 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 17 |
Hb_001307_230 |
0.0299104678 |
- |
- |
PREDICTED: interferon-related developmental regulator 1-like [Jatropha curcas] |
| 18 |
Hb_026317_020 |
0.0311490906 |
- |
- |
hypothetical protein VITISV_035911 [Vitis vinifera] |
| 19 |
Hb_003868_010 |
0.0316683521 |
- |
- |
- |
| 20 |
Hb_001782_090 |
0.0327215539 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g62910-like [Vitis vinifera] |